AMS / ADF
AMS / ADF
General Info
ADF - General Info - Main fields
| Field | Source | Sample value |
|---|---|---|
| Title | Set on Browse calculation publication | Sample calculation |
| Browse Item | URL pointing Browse published item | https://iochem-bd.iciq.es/browse/handle/100/1722 |
| Program | program.header template | ADF 2007 |
| Author | Username fullname | Doe, John |
| Formula | Atom count from final geometry | H 1 O 40 P 1 Ti 1 W 11 |
| Calculation type | Custom logic [^1] | Geometry optimization Minimum |
| Method | DFT | DFT |
ADF - General Info - Additional fields (if thermochemistry module exists)
| Field | Source | Sample value |
|---|---|---|
| Temperature | <scalar dictRef="cc:temp"> | 300 K |
| Pressure | <scalar dictRef="cc:press"> | 1.0 atm |

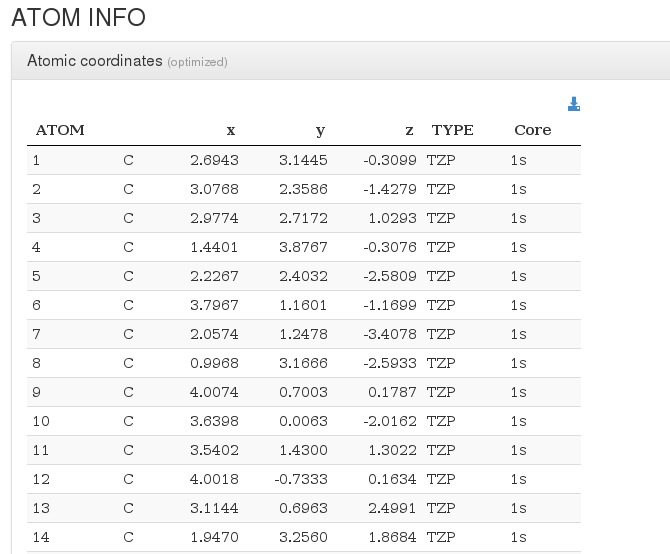
Atoms and Basis Sets
After header section, our HTML resume will output a xyz coordinates table with current molecule atoms.
For every atom, we will output it's serial number, atom type, coordinates in angstroms, basis used and contraction.
In geometry optimizations calculations, next to geometry section header there will appear the word (optimized), pointing that this geometry is the last one from all optimization steps and has converged.
If the geometry optimization did not converge, there will appear the phrase (calculation did not converge).
If there are multiple geometries we'll capture them acording with the following table order :
Atomic coordinates - Possible candidates (most important first)
| Section | Note |
|---|---|
| <module cmlx:templateRef="geometry.cycle"> | Latest geometry cycle(Coordinates on Input orientation) |
| <module cmlx:templateRef="geometry.cycle"> | Lastest geom.opt. (Coordinates) |
| <module cmlx:templateRef="quild.coord"> | Latest quild iteration |
| <module cmlx:templateRef="nuclear.coordinates"> | NMR geometry |
| <module cmlx:templateRef="geometry"> | Single point initial geometry |
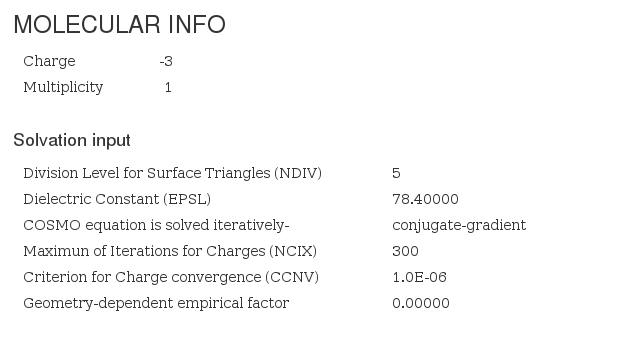
Molecular Info
This section captures molecule additional information not captured on previous section.
Molecular Info - Main fields
| Field | Source | Description |
|---|---|---|
| Spin polarization | <scalar dictRef="a:spinPolarization"> | Variable found inside logfile module (last instance is captured) |
| Multiplicity | Depends on spin polarization | If spin polarization exists, multiplicity = spinpolarization + 1, otherwise multiplicity = 1 |
| Charge | <scalar dictRef="a:charge"> | Variable found inside logfile module (last instance is captured) |
| Solvation | <module cmlx:templateRef="solvation"> | Solvation parameters |
Modules
Bonding energy
Data source: <module cmlx:templateRef='bonding.energy'>

Fit test
Data source: <module cmlx:templateRef='fit.test'>
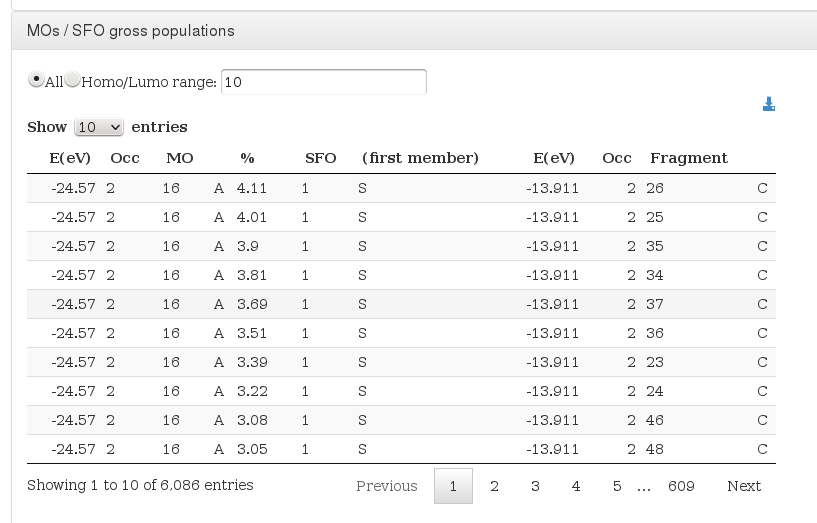
MOs / SFO gross populations
Data source: <module cmlx:templateRef='molecular-orbitals'>
MOs Energies
Data source: <module cmlx:templateRef='orbital.energies'>
Data source: <module cmlx:templateRef='orbital.energies.spin'>
Data source: <module cmlx:templateRef='orbital.energies.spin'>
Mulliken Atomic Charges
Data source <module cmlx:templateRef='mulliken'>
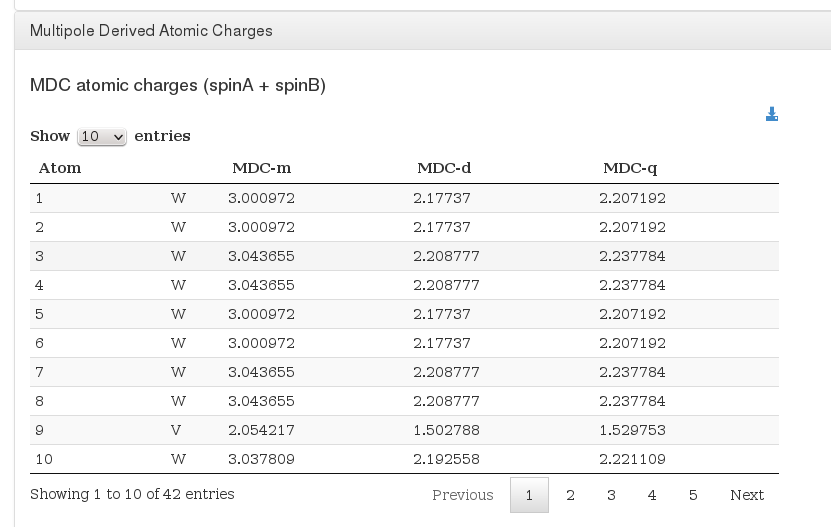
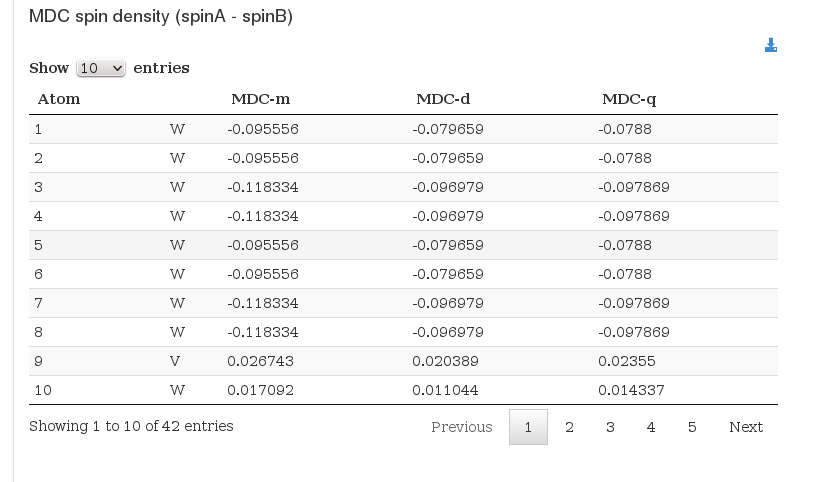
Multipole Derived Atomic Charges
Data source: <module cmlx:templateRef='atomic.charges'>
Data source: <module cmlx:templateRef='atomic.charges.spin'>
Data source: <module cmlx:templateRef='spin.density'>
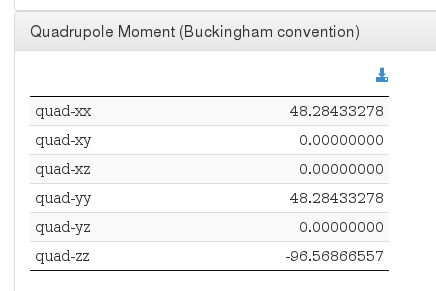
Quadrupole Moment
Data source: <module cmlx:templateRef='quadrupole.moment'>
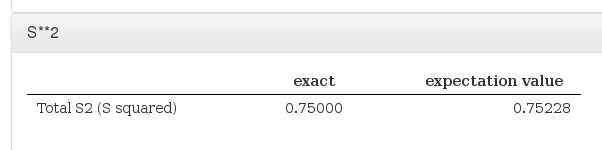
S**2
Data source: <module cmlx:templateRef="s2">
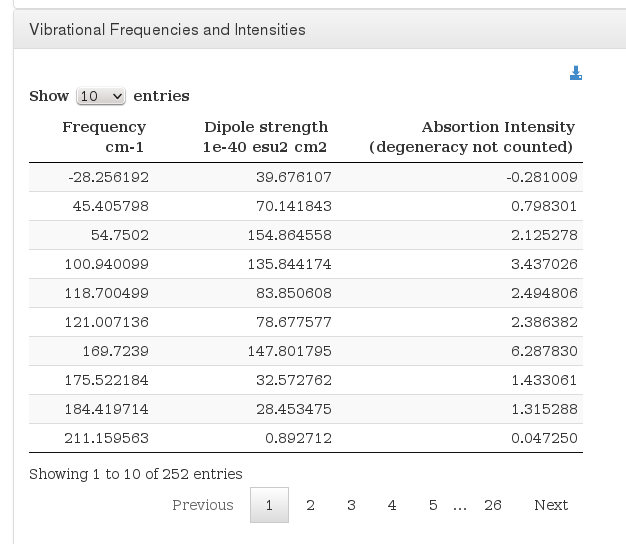
Vibrational Frequencies and Intensities
Data source: <module cmlx:templateRef='intensities'>
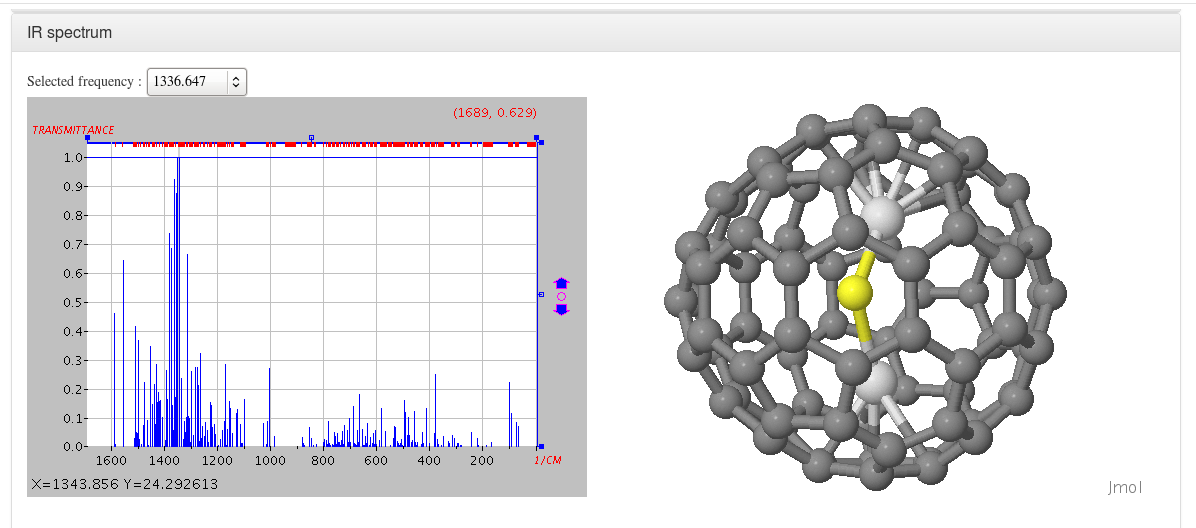
IR spectrum
Data source: <module cmlx:templateRef='vibrations'>
This module will display JSpecView + JSmol plugins (using javascript libraries) working together to represent molecule IR spectrum.
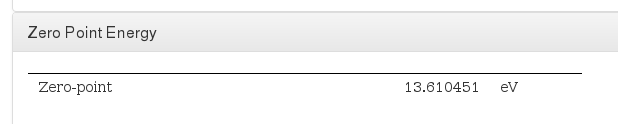
Zero Point Energy
Data source:
- <module cmlx:templateRef="zeropoint"><scalar dictRef="cc:zeropoint">
Thermochemistry
Data source: <module cmlx:templateRef='thermochemistry'>

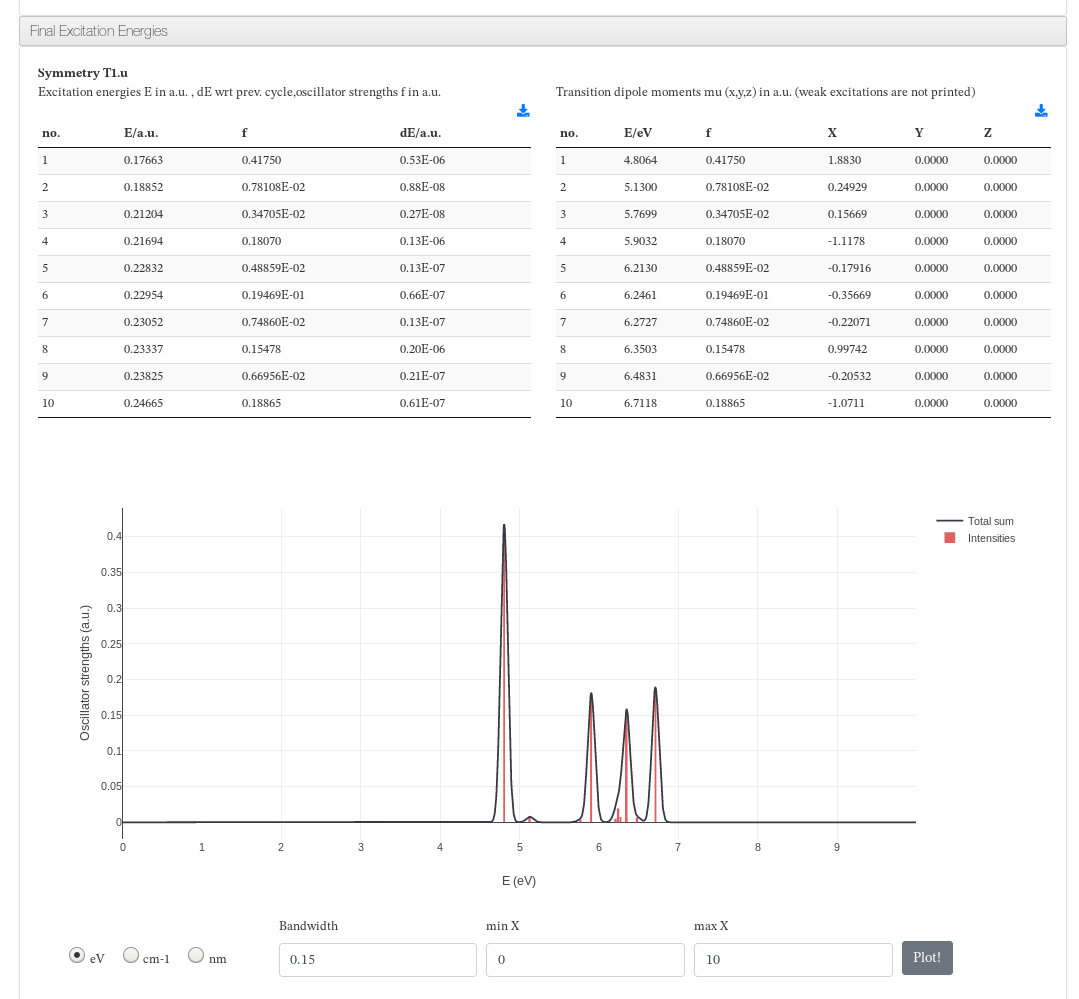
Final Excitation Energies
Data source: <module cmlx:templateRef='excitation.energy'>
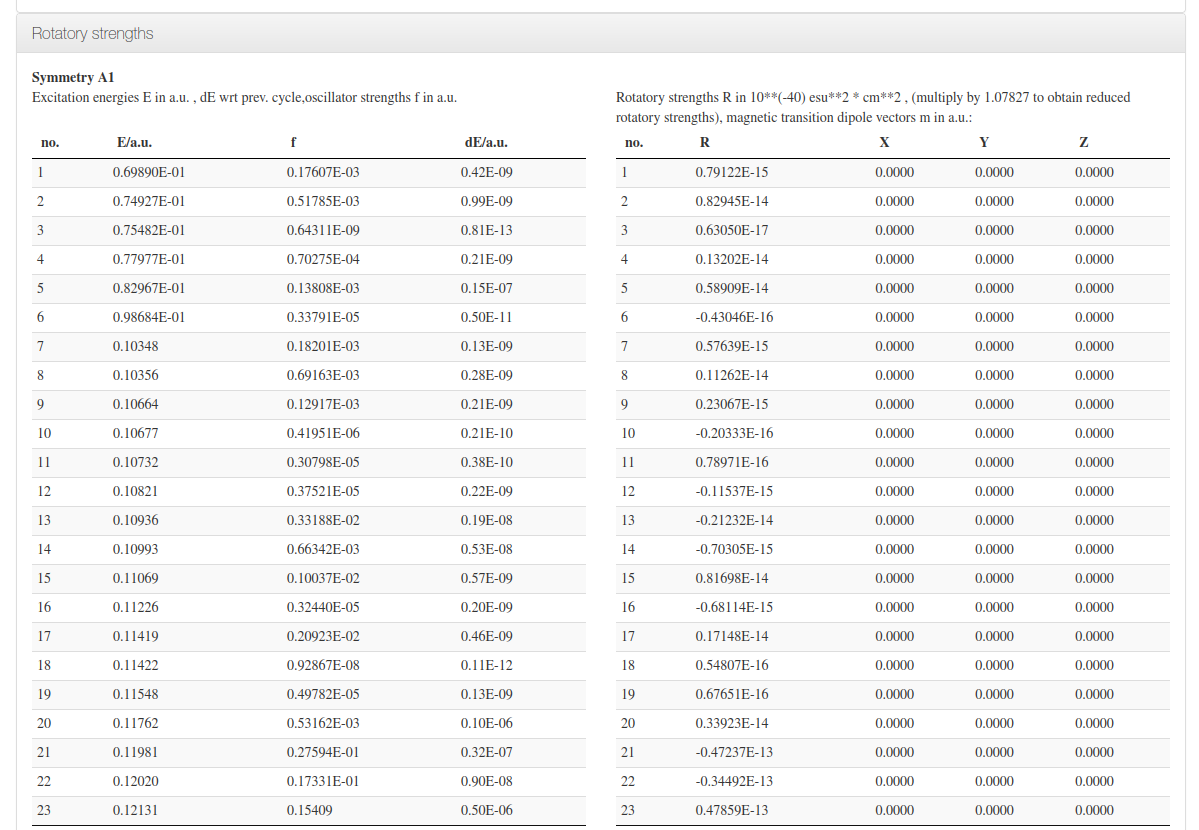
Rotatory Strengths
Data source: <module cmlx:templateRef='excitation.energy'>
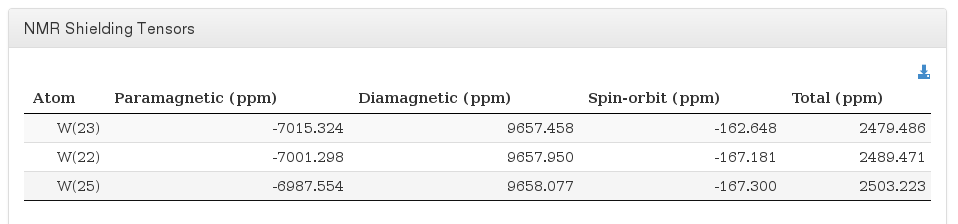
NMR Shielding Tensors
Data source: <module cmlx:templateRef='nmr'>
Timing
Data source: <module cmlx:templateRef='timing'>
Input file
Data source: <module cmlx:templateRef='input.file'>
[^1]: string adf:getCalcType string runtype boolean hasVibrations boolean isMininum boolean isQuild boolean isNMR
$runtype Refers to <scalar dataType="xsd:string" dictRef="cc:runtype">
$hasVibrations Exists module <module cmlx:templateRef="vibrations" > ?
$isMinimum All frequencies from <module cmlx:templateRef="vibrations" > are positive?
$isQuild Exists module <module cmlx:templateRef="quild.iteration" > ?
$isNMR Exists module <module cmlx:templateRef="nucleus" > ?
<!-- Calculation type related constants -->
<xsl:variable name="adf:GeometryOptimization" select="'Geometry optimization'" />
<xsl:variable name="adf:SinglePoint" select="'Single point'" />
<xsl:variable name="adf:TransitionState" select="'TS'" />
<xsl:variable name="adf:Frequencies" select="'Frequencies'" />
<xsl:variable name="adf:Minimum" select="'Minimum'"/>
<xsl:variable name="adf:Quild" select="'Quild'" />
<xsl:variable name="adf:NMR" select="'NMR'" />
<!-- Calculation type variables -->
<xsl:variable name="calcType" select="
if(compare($runType,'GEOMETRY OPTIMIZATION') = 0)
then $adf:GeometryOptimization
else
if(compare($runType,'SINGLE POINT') = 0)
then $adf:SinglePoint
else
if(compare($runType,'TRANSITION STATE') = 0)
then $adf:TransitionState
else
if(compare($runType,'FREQUENCIES') = 0)
then $adf:Frequencies
else
$adf:SinglePoint" />
<xsl:variable name="vibrations" select="
if($hasVibrations)
then if($isMinimum)
then concat(' ', $adf:Minimum)
else
if(compare($calcType,$adf:TransitionState) != 0)
then concat(' ',$adf:TransitionState)
else
''
else ''" />
<xsl:variable name="quild" select="
if($isQuild)
then concat(' ',$adf:Quild)
else
''" />
<xsl:variable name="nmr" select="
if($isNMR)
then concat(' ',$adf:NMR)
else
''"
/>
<xsl:sequence select="concat($calcType, $vibrations, $quild, $nmr)"/>