VASP
Vasp
General Info
VASP - General Info - Main fields
| Field | Source | Sample value |
|---|---|---|
| Title | Set on Browse calculation publication | Sample calculation |
| Browse Item | URL pointing Browse published item | https://iochem-bd.iciq.es/browse/handle/100/1722 |
| Program | <scalar dictRef="cc:program"> template | VASP 5.3.2 |
| Author | Username fullname | Doe, John |
| Formula | Atom count from final geometry | C 1 H 4 Mo 30 O 61 |
| Calculation type | Custom logic [^1] | Geometry optimization |
| Functional | Custom logic [^2] | PBE+U |
| Shell type (ISPIN) | <scalar dictRef="v:ispin"> | Open Shell |
| Temperature | <scalar dictRef="cc:temp"> | 0.0K |

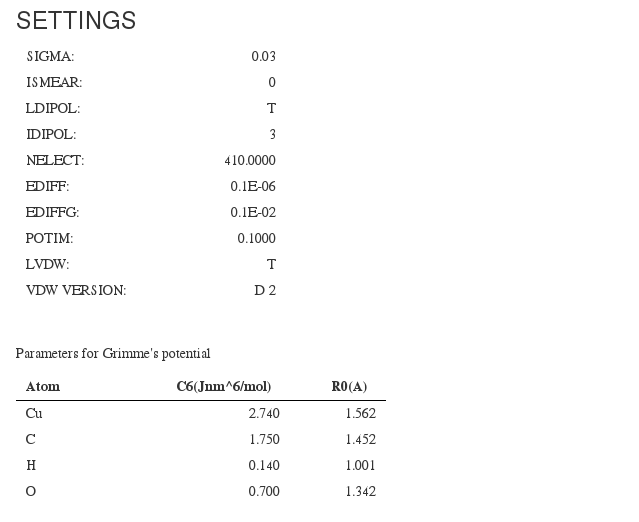
Settings
Most relevant calculation input parameters. Almost all information fields come from <module cmlx:templateRef="incar">
SIGMA
ISMEAR
LDIPOL
IDIPOL
Multiplicity
NELECT
Cut-Off Energy
EDIFF
EDDIFG
POTIM
LDAU (LDAUL, LDAUU, LDAUJ)
VDW and VDW Version
Parameters for Grimme's potential <module cmlx:templateRef="grimmes">
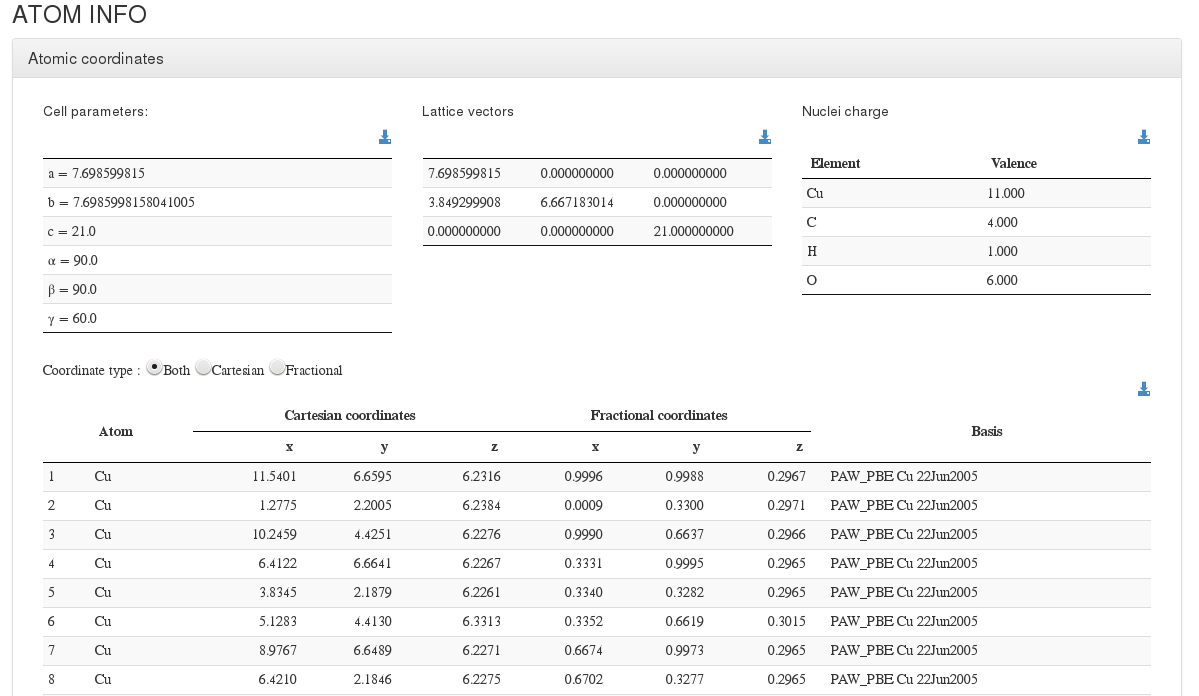
Atoms info
After settings section, our HTML resume will output cell coordinates and a coordinates table with molecule atoms.
Initial geometry its read from OUTCAR file using: <module cmlx:templateRef="position">, <module cmlx:templateRef="incar">, <module cmlx:templateRef="potcar"> and <module cmlx:templateRef="laticce">
Final geometry will be generated using the same modules than Initial geometry, but coordinates will come from last instance of <module cmlx:templateRef="calculated.position">
NEB and DIM calculation types will display buttons to choose the displayed geometry.
On geometry optimization calculations, the initial and final geometries will be displayed side to side on a JSmol viewer.
For every atom, we will output it's serial number, atom type, cartesian and fractional coordinates (in angstroms) , and basis used.
Molecular Info
This section captures molecule additional information not captured on previous section.
K-point generation parameters read from KPOINTS file

Modules
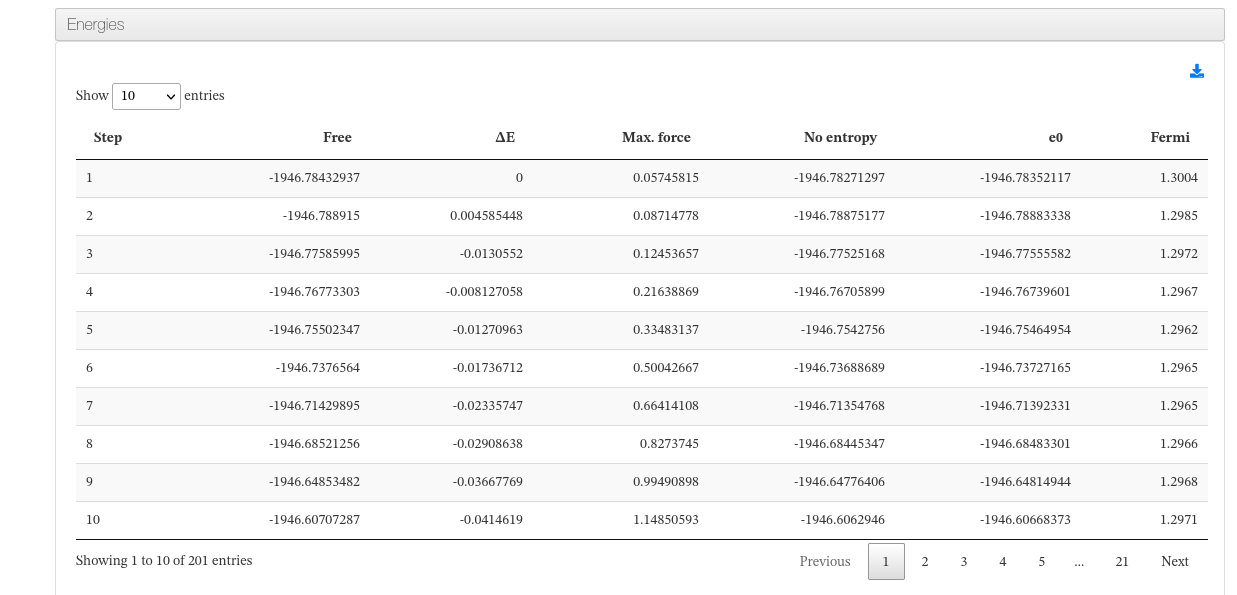
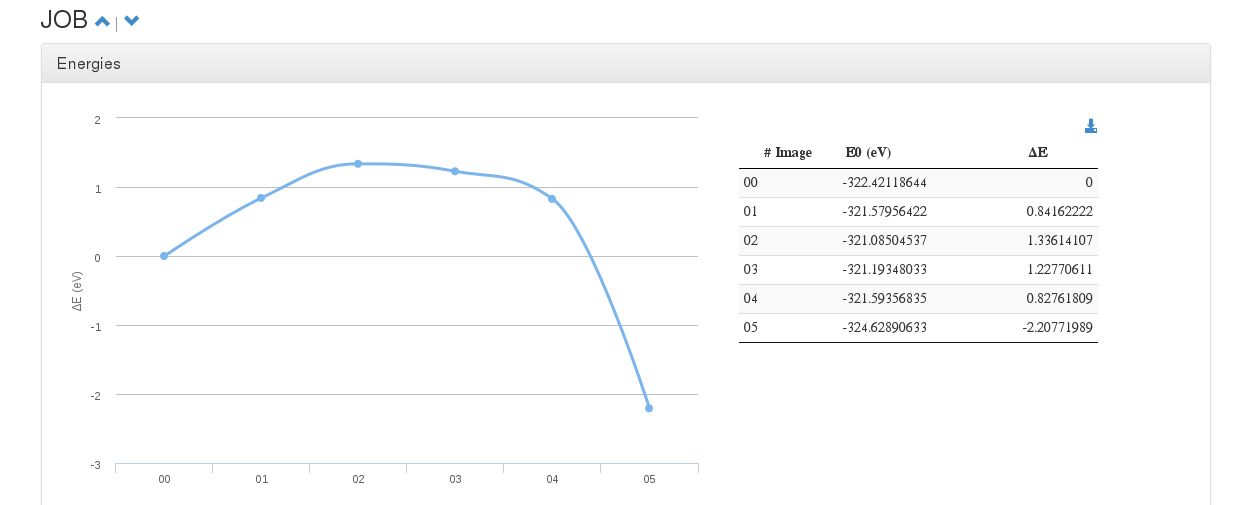
Energies
Data source: <module cmlx:templateRef='energy'>
This module will hold Free ,E0 , dE and E-fermi energies.
In case of single calculations (single point, geometry optimization), a table with this values will be displayed.

On Molecular Dynamics calculations, a table with energy and force progression will appear along with a plot of such values.
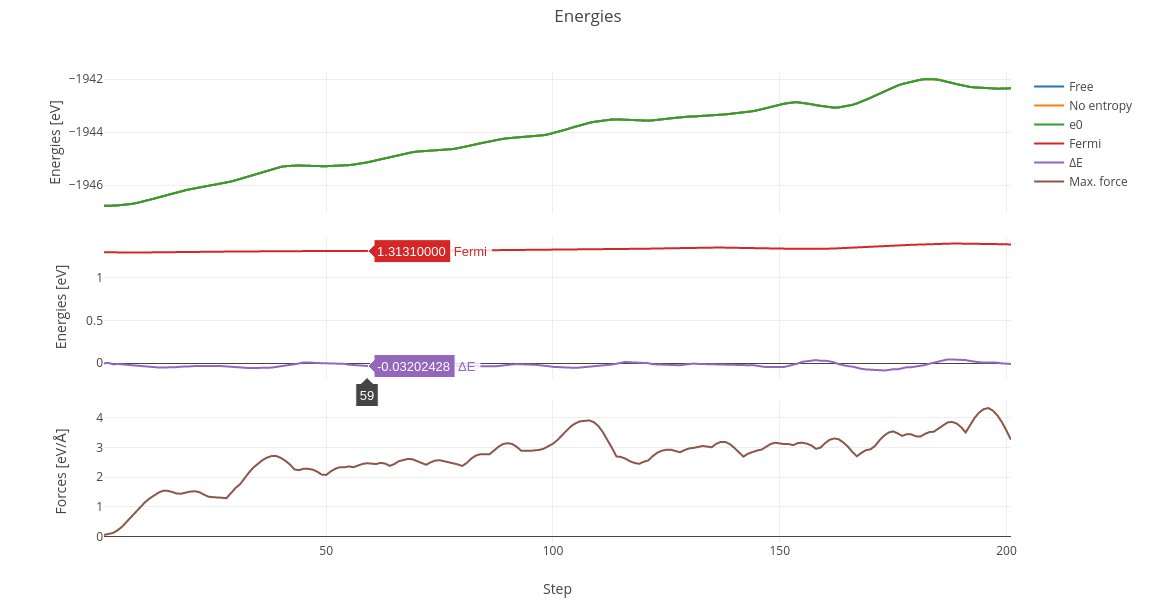
On multiple OUTCAR calculations like Nudge Elastic Band (NEB) or Dimmer, a graphic will be printed with each diferential energy as step.
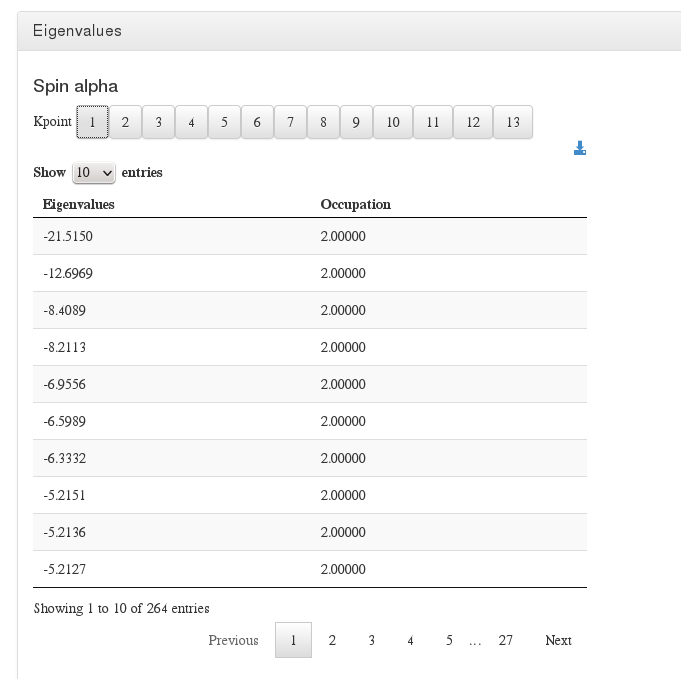
Eigenvalues
Data source: <module cmlx:templateRef='eigenvalues'>
This module will display eigenvalues per spin and kpoint.
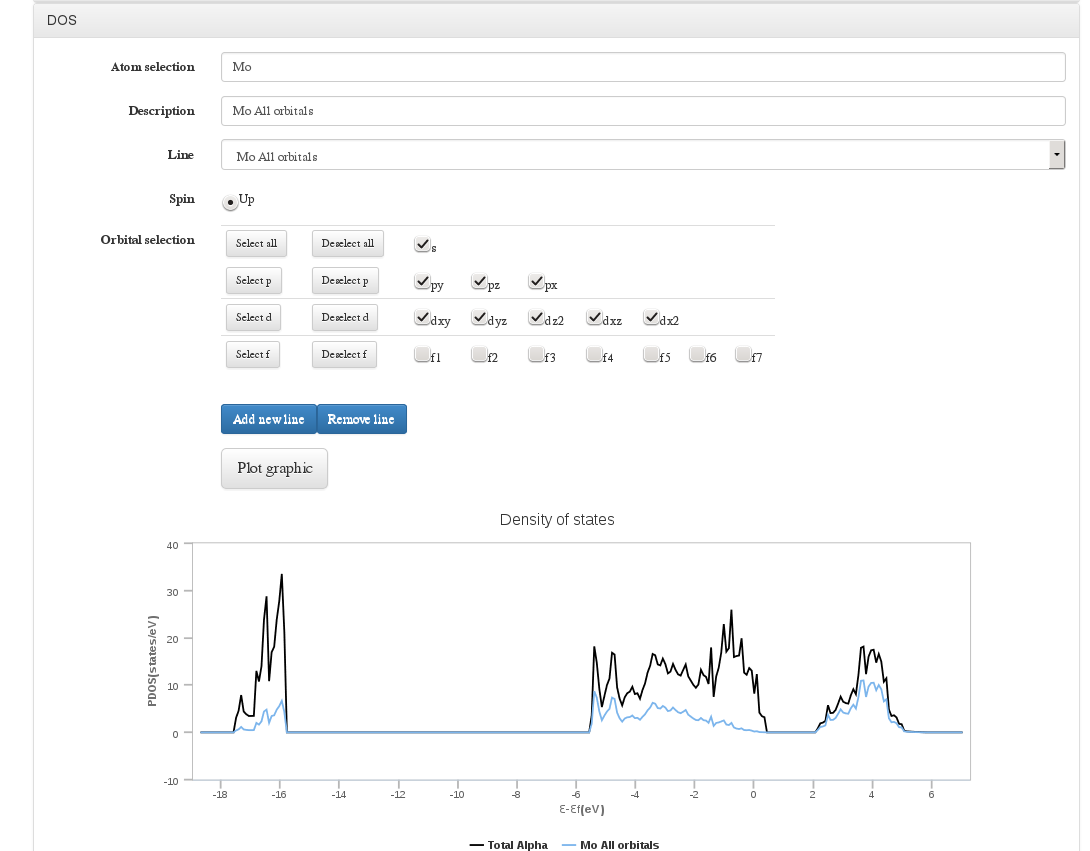
DOS
Data source: DOSCAR file <module cmlx:templateRef='vasp.doscar'>
On calculations where VASP DOSCAR file has been uploaded, a form will be displayed to configure a graph with the Density Of States (DOS) information
In this form we will can select atoms by index, range or atom type, select spin and molecular orbitals. Once all parameters have been set we will add this line to current graph. We can define/delete as much DOS lines as we need.
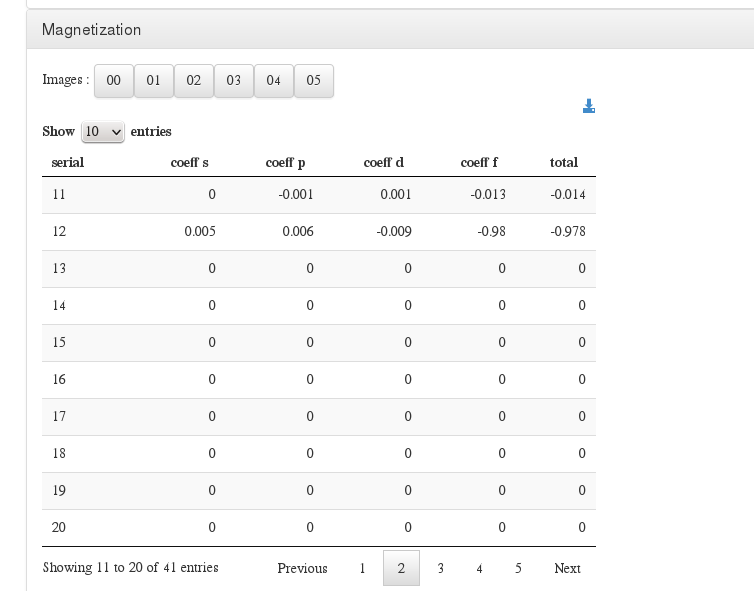
Magnetization
Data source: <module cmlx:templateRef='magnetization'>
Vibrations
Data source: <module cmlx:templateRef='vibrations'>
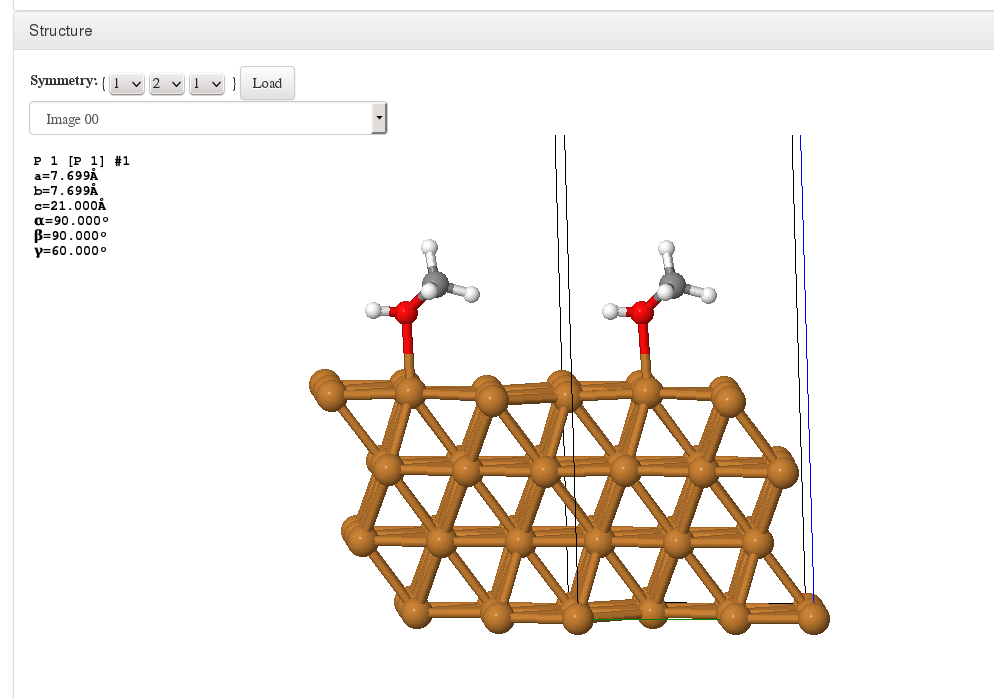
Structure
Lattice replication
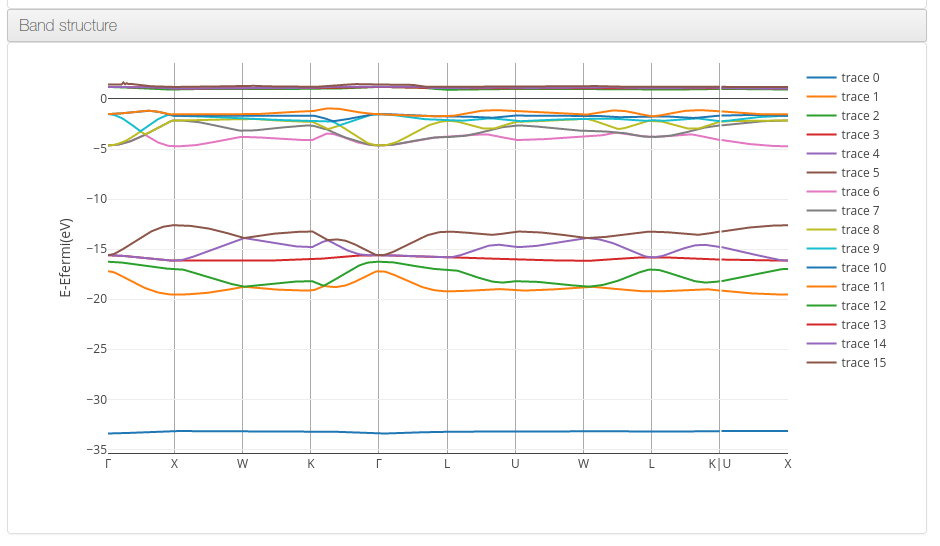
Band structure
Data source: <module id='kpoints'> and <module cmlx:templateRef='eigenvalues'>
On Band structure calculations the platform will display a graph with the eigenvalues for each band, centering zero on the Fermi energy value.
[^1]: string vasp:getCalcType nodeset ibrion
ibrion Value of ibrion parameter on <module cmlx:templateRef="convergence.info" >.
<xsl:param name="ibrion"/>
<xsl:choose>
<xsl:when test="count($ibrion) > 1 and exists($ibrion[text() = '44'])"><xsl:value-of select="$vasp:ImprovedDimerMethod"/></xsl:when>
<xsl:when test="count($ibrion) > 1"><xsl:value-of select="$vasp:NudgedElasticBand"/></xsl:when>
<xsl:otherwise>
<xsl:choose>
<xsl:when test="$ibrion = -1"><xsl:value-of select="$vasp:SinglePoint"/></xsl:when>
<xsl:when test="$ibrion = 0"><xsl:value-of select="$vasp:MolecularDynamics"/></xsl:when>
<xsl:when test="$ibrion > 0 and $ibrion < 4"><xsl:value-of select="$vasp:GeometryOptimization"/></xsl:when>
<xsl:when test="$ibrion > 4 and $ibrion < 9"><xsl:value-of select="$vasp:FrequencyCalculus"/></xsl:when>
<xsl:when test="$ibrion = 44"><xsl:value-of select="$vasp:ImprovedDimerMethod"/></xsl:when>
<xsl:otherwise><xsl:value-of select="$vasp:NotAvailable"/></xsl:otherwise>
</xsl:choose>
</xsl:otherwise>
</xsl:choose>[^2]: string turbo:getMehod string gga boolean lhfcalc number hfscreen number aggac boolean luseVdw number zabVdw number param1> number param2 boolean ldau
gga, lhfcalc, hfscreen , ... parameters read from OUTCAR file <module cmlx:templateRef="incar">
<xsl:param name="gga"/>
<xsl:param name="lhfcalc"/>
<xsl:param name="aggac"/>
<xsl:param name="hfscreen"/>
<xsl:param name="luseVdw"/>
<xsl:param name="zabVdw"/>
<xsl:param name="param1"/>
<xsl:param name="param2"/>
<xsl:param name="ldau"/>
<xsl:choose>
<xsl:when test="compare($lhfcalc,'true')=0">
<xsl:choose>
<xsl:when test="$hfscreen=0.2">HSE06</xsl:when>
<xsl:otherwise>HSE03</xsl:otherwise>
</xsl:choose>
</xsl:when>
<xsl:when test="compare($luseVdw , 'true')=0 and $aggac = 0.0">
<xsl:choose>
<xsl:when test="compare($gga,'RE')=0">vdW-DF</xsl:when>
<xsl:when test="compare($gga,'OR')=0">optPBE-vdW</xsl:when>
<xsl:when test="compare($gga,'BO')=0 and round($param1 * 1000) div 1000 = 0.183 and round($param2 * 100) div 100 = 0.22">optB88-vdW</xsl:when>
<xsl:when test="compare($gga,'MK')=0 and round($param1 * 10000) div 10000 = 0.1234 and $param2 = 1.0">optB86d-vdW</xsl:when>
<xsl:when test="compare($gga,'ML')=0 and $zabVdw = -1.8867">vdW-DF2</xsl:when>
<xsl:otherwise>N/A</xsl:otherwise>
</xsl:choose>
</xsl:when>
<xsl:when test="compare($gga,'91')=0">PW91</xsl:when>
<xsl:when test="compare($gga,'PE')=0">PBE</xsl:when>
<xsl:when test="compare($gga,'RP')=0">rPBE</xsl:when>
<xsl:when test="compare($gga,'AM')=0">AM05</xsl:when>
<xsl:when test="compare($gga,'PS')=0">PBEsol</xsl:when>
<xsl:otherwise>N/A</xsl:otherwise>
</xsl:choose>
<xsl:if test="compare($ldau,'true')=0">
<xsl:text>+U</xsl:text>
</xsl:if>