Turbomole
Turbomole
General Info
Turbomole - General Info - Main fields
| Field | Source | Sample value |
|---|---|---|
| Title | Set on Browse calculation publication | Sample calculation |
| Browse Item | URL pointing Browse published item | https://iochem-bd.iciq.es/browse/handle/100/1722 |
| Program | <scalar dictRef="cc:program"> template | Turbomole 5.3.2 |
| Author | Username fullname | Doe, John |
| Formula | Atom count from final geometry | C 6 H 12 Fe 1 N 24 |
| Calculation type | Custom logic [^1] | Geometry optimization Minimum |
| Method(s) | Custom logic [^2] | DFT (b3-lyp, D3, ri-j, gridsize:m3) |
Turbomole - General Info with additional fields (if freeh module exists)
| Field | Source | Sample value |
|---|---|---|
| Temperature | <scalar dictRef="cc:temp"> | 298.15 K |
| Pressure | <scalar dictRef="cc:press"> | 1.0 atm |


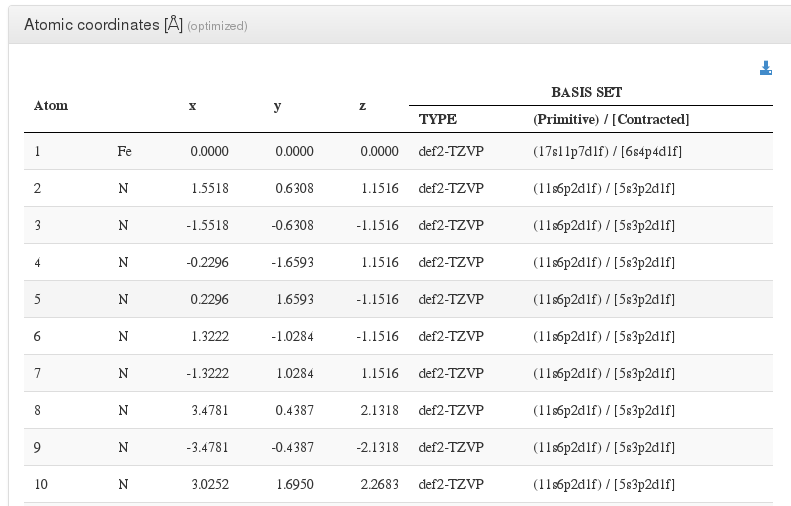
Atoms and Basis Sets
After header section, our HTML resume will output a xyz coordinates table with current molecule atoms.
Initially its read from coord file
Then we will read all instances from module <module cmlx:templateRef="atomcoord"> and use last instance as final geometry
For every atom, we will output it's serial number, atom type, coordinates in angstroms, and basis used.
In geometry optimizations calculations, next to geometry section header there will appear the word (optimized), pointing that this geometry is the last one from all optimization steps and has converged.
If the geometry optimization did not converge, there will appear the phrase (calculation did not converge).
If there are multiple geometries we'll capture it's last appearance.
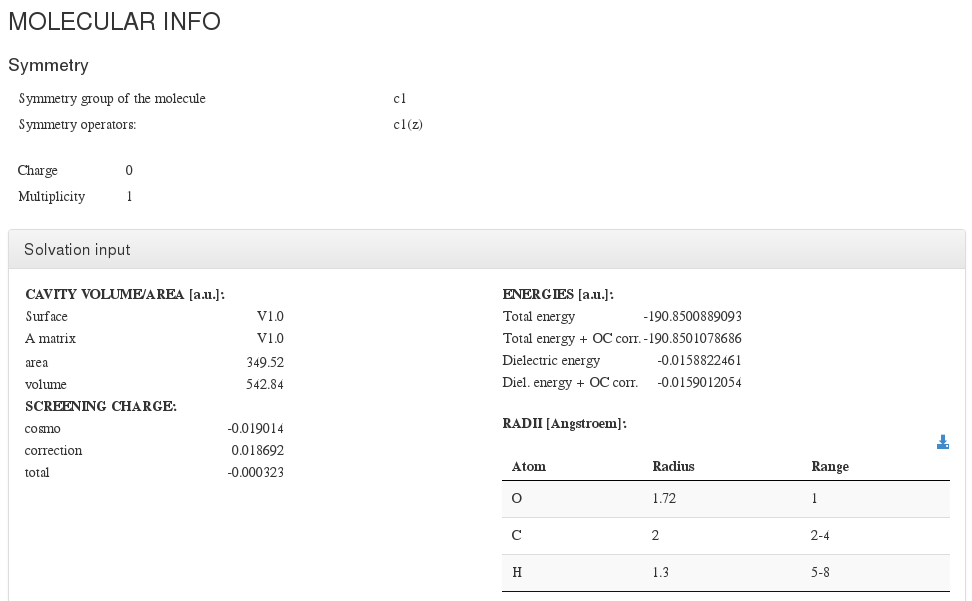
Molecular Info
This section captures molecule additional information not captured on previous section.
Molecular Info - Main fields
| Field | Source | Description |
|---|---|---|
| Symmetry | Appears in symmetry module. | Symmetry information about the molecule (if it exists) |
| Multiplicity | Its value is set:
| |
| Charge | Readed from:
| |
| Geometry restrictions | <module cmlx:templateRef="restrictions"> | User defined geometry restrictions |
| Solvation | <module cmlx:templateRef="cosmo"> | Solvation parameters |
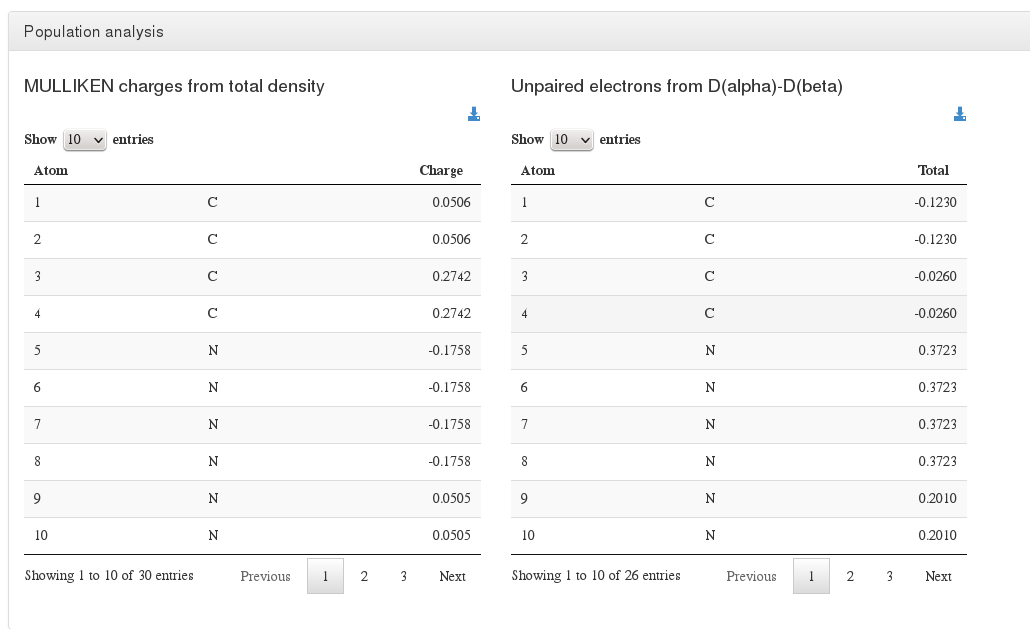
Modules
Population analysis
Data source: <module cmlx:templateRef='population.analysis'>
Data source: <module cmlx:templateRef='fit.pointcharges'>
This module will hold Mulliken, Loewdin and Natural population analysis, will also contain (if exists) information from unpaired electrons from D(alpha)-D(beta)
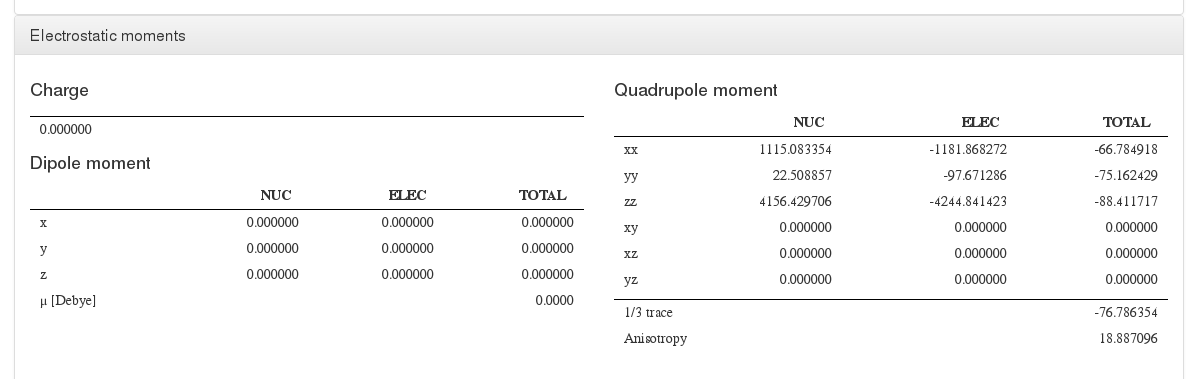
Electrostatic moments
Data source: <module cmlx:templateRef='electrostatic.moments'>
This module will display charge, dipole and multipole values.
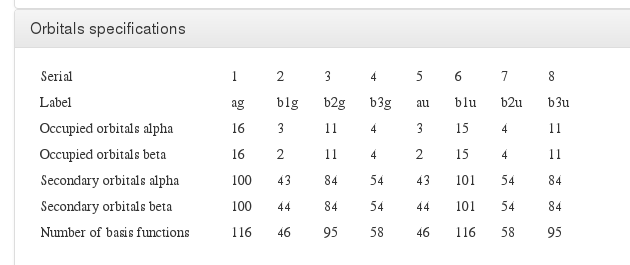
Orbital specification
Data source: <module cmlx:templateRef='orbitals'>
Final results
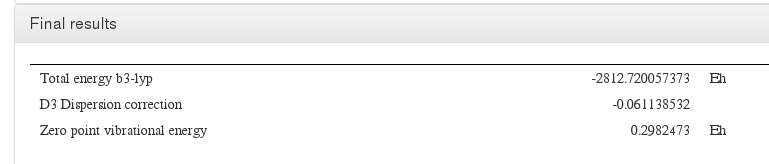
Data source: <module cmlx:templateRef='turbomole.energy'> Taken from last line of energy file
Data source: <module cmlx:templateRef='energy'>
Data source: <module cmlx:templateRef='nuclear.repulsion'>
Data source: <module cmlx:templateRef='zero.point.energy'>
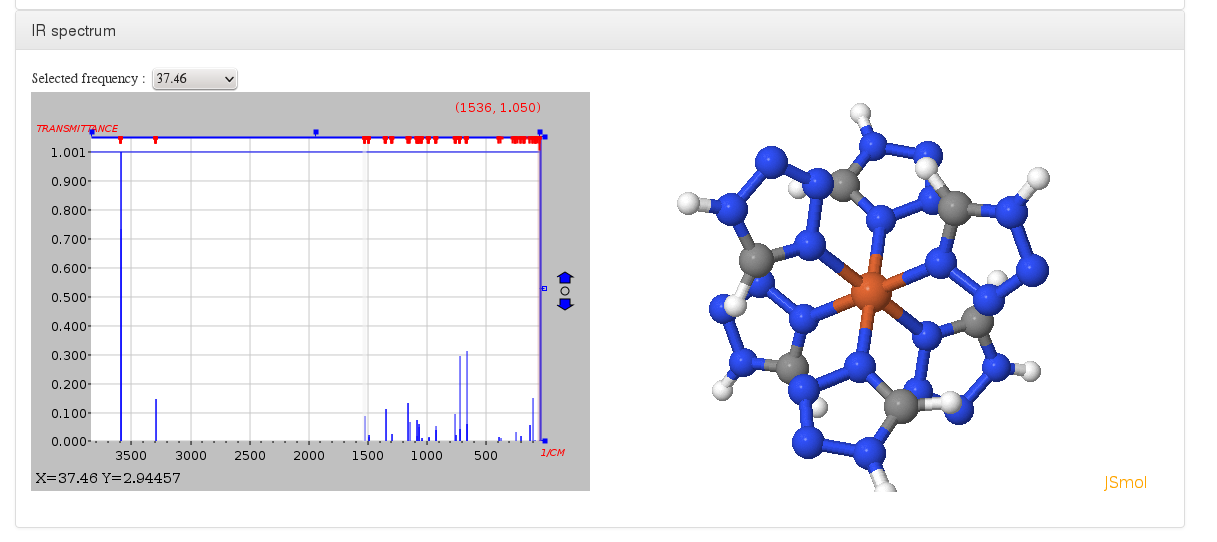
IR spectrum
Data source: <module cmlx:templateRef='vibrations'>
This module will display JSpecView + JSmol plugins (using javascript libraries) working together to represent molecule IR spectrum.
All information will come from "$vibrational normal modes" and "$vibrational spectrum" sections inside Turbomole control file, in case they are defined on external files such as vib_normal_modes and vibspectrum we must copy them inside control file.
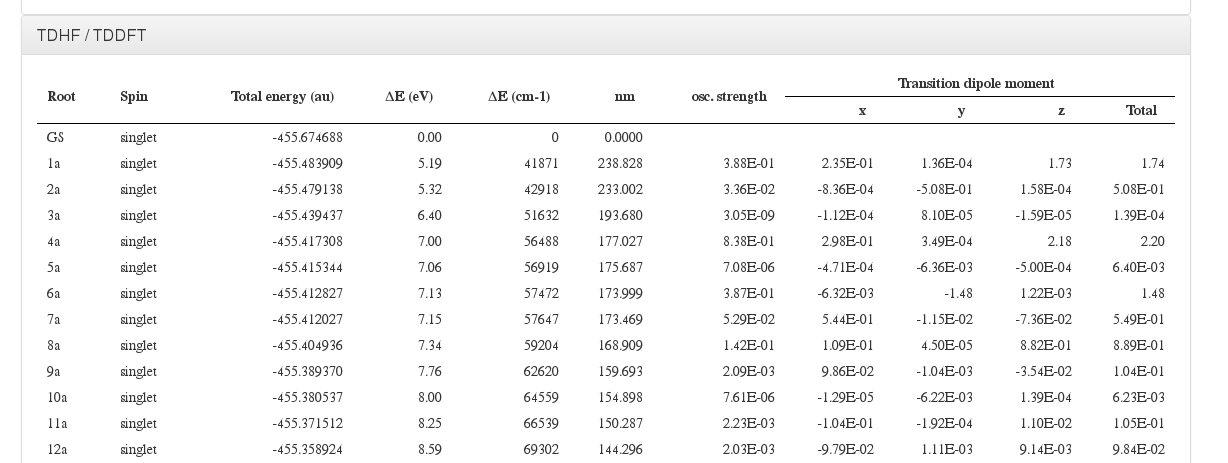
TDDFT/TDHF
Data source: <module cmlx:templateRef='excitation'>
[^1]: string turbo:getCalcType boolean isRestrictedOptimization boolean isOptimization boolean isIncomplete nodeset vibrations nodeset statpt nodeset soes
$isRestrictedOptimization Exists module <module cmlx:templateRef="restrictions" > ?
$isOptimization Exists module <module cmlx:templateRef="convergence.info" > ?
$isIncomplete Last module <module cmlx:templateRef="convergence.info" > has a "NO" on converged fields?
$vibrations Vibrational frequencies information, headers and displacements. Refer to <module cmlx:templateRef="vibrations" >
$statpt statpt parameters section read from control file
$soes soes parameters section read from control file
<xsl:param name="isRestrictedOptimization" as="xs:boolean"/>
<xsl:param name="isOptimization" as="xs:boolean"/>
<xsl:param name="isIncomplete" as="xs:boolean"/>
<xsl:param name="vibrations" as="node()?"/>
<xsl:param name="statpt" as="node()?"/>
<xsl:param name="soes" as="node()?"/>
<xsl:variable name="isMinimum" select="not(contains(replace($vibrations/cml:module[@cmlx:templateRef='spectrum']/array[@dictRef='cc:frequency'],'-0.00',''), '-'))"/>
<xsl:variable name="isExcitedState">
<xsl:if test="exists($soes) and number($soes/cml:array[@dictRef='t:irrep']/@size) = 1">
<xsl:value-of select="$turbo:ExcitedState"/>
<xsl:text> </xsl:text>
<xsl:if test="$isRestrictedOptimization or $isOptimization">
(<xsl:value-of select="$soes/cml:array[@dictRef='t:lowest']"/><xsl:value-of select="$soes/cml:array[@dictRef='t:irrep']"/>)
</xsl:if>
</xsl:if>
</xsl:variable>
<xsl:variable name="isTS" select="
if(exists($statpt) and number($statpt//cml:scalar[@dictRef='t:itrvec']) > 0) then
$turbo:TransitionState
else
''
"/>
<xsl:variable name="itrvecdsd" select="number($statpt//cml:scalar[@dictRef='t:itrvec'])"/>
<xsl:variable name="calcType" select="
if($isRestrictedOptimization) then
$turbo:RestrictedGeometryOptimization
else if($isOptimization) then
concat($turbo:GeometryOptimization, ' ', $isTS)
else
$turbo:SinglePoint
"/>
<xsl:variable name="vibration" select="
if(exists($vibrations) and not($isRestrictedOptimization) and compare($isTS,'') = 0) then
if($isMinimum) then
$turbo:Minimum
else
$turbo:TransitionState
else ''
"/>
<xsl:sequence select="concat($calcType, ' ', $vibration, ' ', $isExcitedState)"/>[^2]: string turbo:getMehod nodeset soes nodeset methodScalar
$soes soes parameters section read from control file <module cmlx:templateRef="soes">
$methodScalar methods read from control file ($dft|$uhf) <module cmlx:templateRef="methods">
<xsl:variable name="methodsTmp">
<xsl:for-each select="$methodScalar">
<xsl:for-each select="tokenize(.,'\s+')">
<xsl:element name="method">
<xsl:value-of select="upper-case(.)"/>
<xsl:text> </xsl:text>
</xsl:element>
</xsl:for-each>
</xsl:for-each>
</xsl:variable>
<xsl:choose>
<xsl:when test="not(exists($methodScalar))">
<xsl:sequence select="
if(exists($soes)) then
'TDHF'
else
'HF'
">
</xsl:sequence>
</xsl:when>
<xsl:otherwise>
<xsl:variable name="step1">
<xsl:choose>
<xsl:when test="contains($methodsTmp, 'RIR12') and contains($methodsTmp, 'MP2') and contains($methodsTmp, 'RICC2')">
<xsl:text>MP2-F12 </xsl:text>
<xsl:value-of select="replace(replace(replace($methodsTmp,'RIR12', ''), 'MP2', ''), 'RICC2', '')"></xsl:value-of>
</xsl:when>
<xsl:otherwise><xsl:value-of select="$methodsTmp"/></xsl:otherwise>
</xsl:choose>
</xsl:variable>
<xsl:variable name="step2">
<xsl:choose>
<xsl:when test="contains($step1, 'UHF') and contains($step1, 'DFT')">
<xsl:text>U-DFT </xsl:text>
<xsl:value-of select="replace(replace($step1,'UHF',''), 'DFT','')"/>
</xsl:when>
<xsl:otherwise><xsl:value-of select="$step1"/></xsl:otherwise>
</xsl:choose>
</xsl:variable>
<xsl:variable name="step3">
<xsl:choose>
<xsl:when test="contains($step2, 'DFT') and exists($soes)">
<xsl:value-of select="replace($step2,'DFT', 'TDDFT')"/>
</xsl:when>
<xsl:otherwise><xsl:value-of select="$step2"/></xsl:otherwise>
</xsl:choose>
</xsl:variable>
<xsl:value-of select="$step3"/>
</xsl:otherwise>
</xsl:choose>