LAMMPS
LAMMPS
For this specific format, the minimum required files are:
A user defined input file.
A data file with the initial atomic cooordinates and cell dimensions, used with the read_data command.
The output file generated by LAMMPS, named log.lammps by default.
A (g)zipped file with all the generated trajectories, or in its absence, a single file with all the trajectories in it, created with the dump command.
NOTE
LAMMPS allows to run the read_data command multiple times to compose the final atoms of the system. ioChem-BD only supports one data file to be uploaded to the platform. Additionals files used to compose the final geometry with the commands read_restart and create_atoms are currently not supported.
Almost all of the atom_styles are supported except hybrid and tdpd.
All units parameter values are supported and the report will display units accordingly.
The platform will render the trajectory files in the HTML report and also keep them inside the platform for its later download. Additionally will derive from them a file called multistep.cif holding all the steps in CIF format.
General Info
LAMMPS - General Info - Main fields
| Field | Source | Sample value |
|---|---|---|
| Title | Set on Browse calculation publication | Sample calculation |
| Browse Item | URL pointing Browse published item | https://iochem-bd.iciq.es/browse/handle/100/5672 |
| Program | program.header template | LAMMPS 29 Oct 2020 |
| Author | Username fullname | Doe, John |
| Calculation type | Custom logic [^1] [^2] | Molecular Dynamics (NPT) (T=Nose-Hoover, P=Nose-Hoover) |

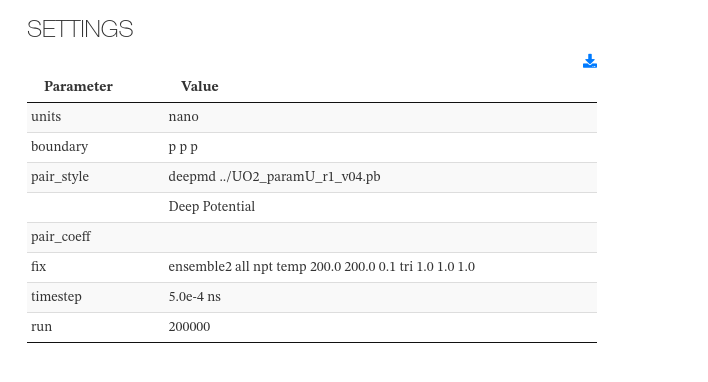
Settings
This section captures initialization settings like units used, potentials, boundaries style, time, timestep used, etc.
Data source: <module cmlx:templateRef='setup'>
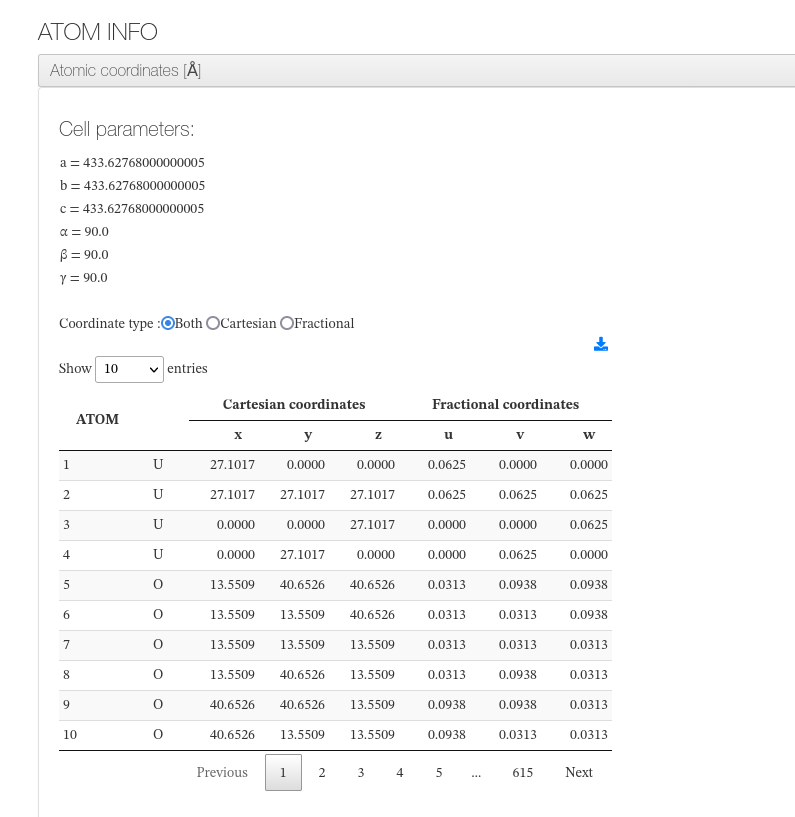
Atomic coordinates
After settings, our HTML resume will output cell dimensions and an xyz coordinates table with current molecule atoms.
For every atom, we will output it's serial number, atom type, coordinates in angstroms in cartesian and fractional types.
Information its read from read_data structure file and parsed based on the atom_style defined.
Modules
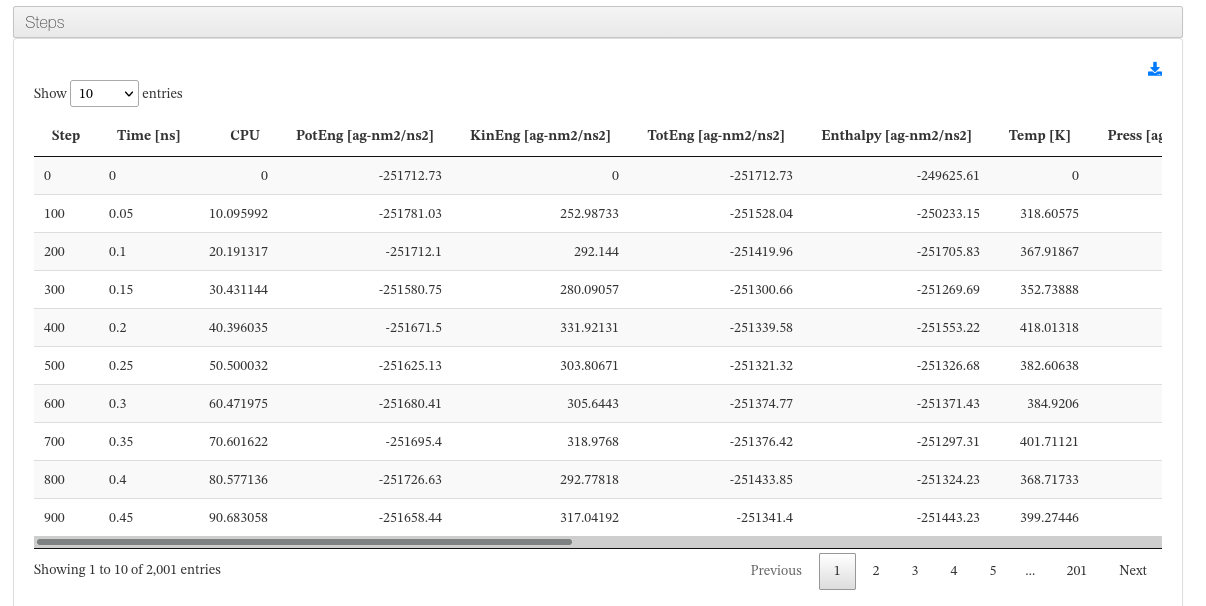
Step resume
Data source: <module cmlx:templateRef='steps'>
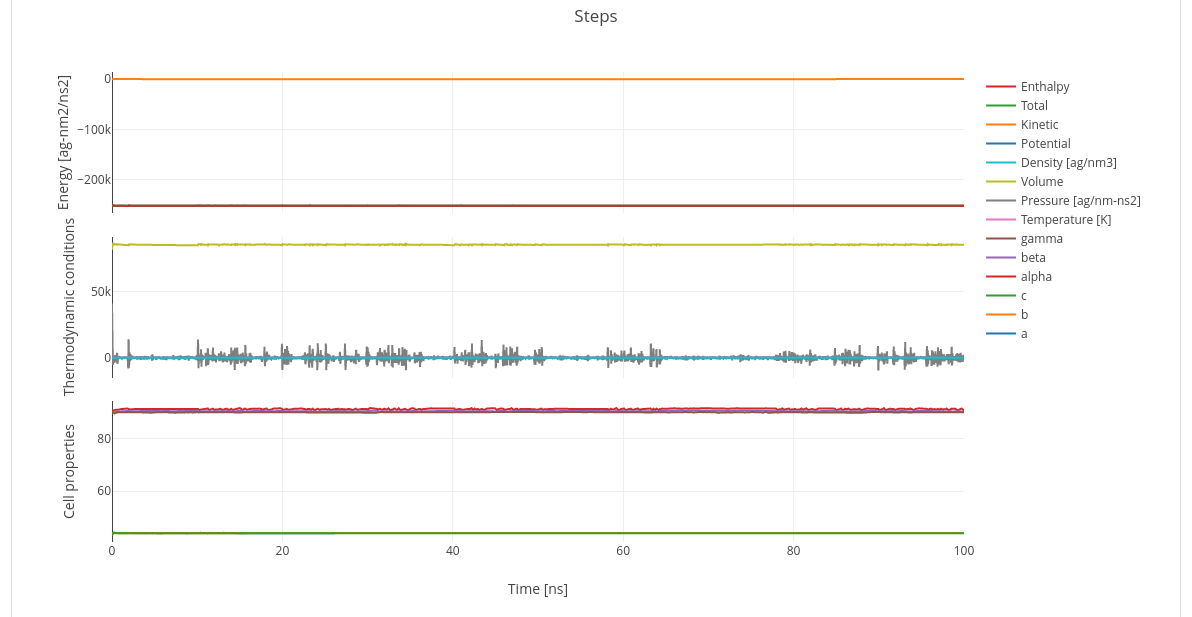
This section is composed of a summary table with the energies for every timestep along with the thermodyamic conditions (pressure and temperature) and cell dimension evolution (length and angles).
Next to it a dynamic graph will display those table values, grouping them by type.
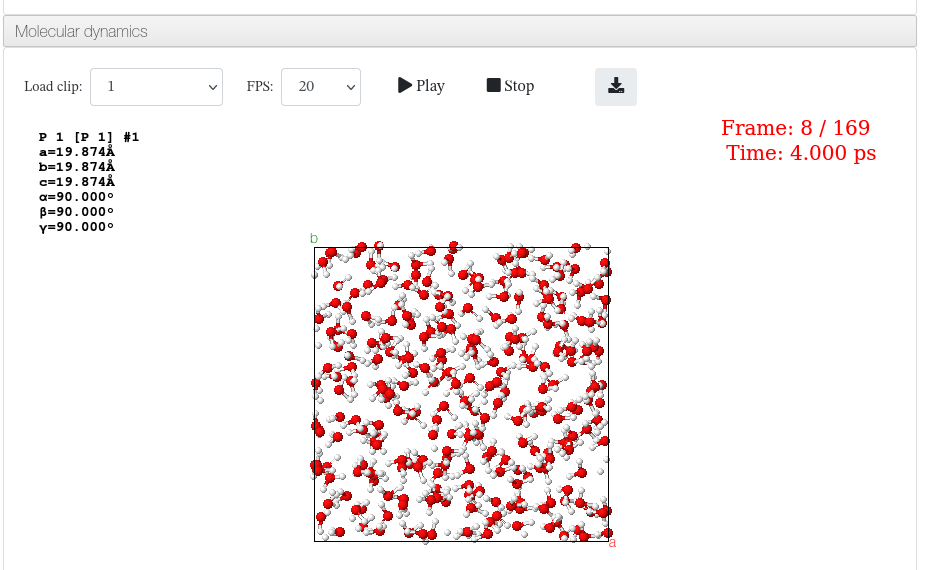
Molecular dynamics
Data source: Trajectory steps extracted from the original LAMMPS trajectory file.
The provided steps are uncompressed, converted into CIF format and concatenated into a single file called *multistep.cif*.
To render it, users will select the desired clip and click the Play button to load it.
The number of rendered images per clip will be based on the size of the contained molecule. On large molecules (+1000 atoms), the animation will render 50 steps per clip, otherwise there will be 200 steps per clip.
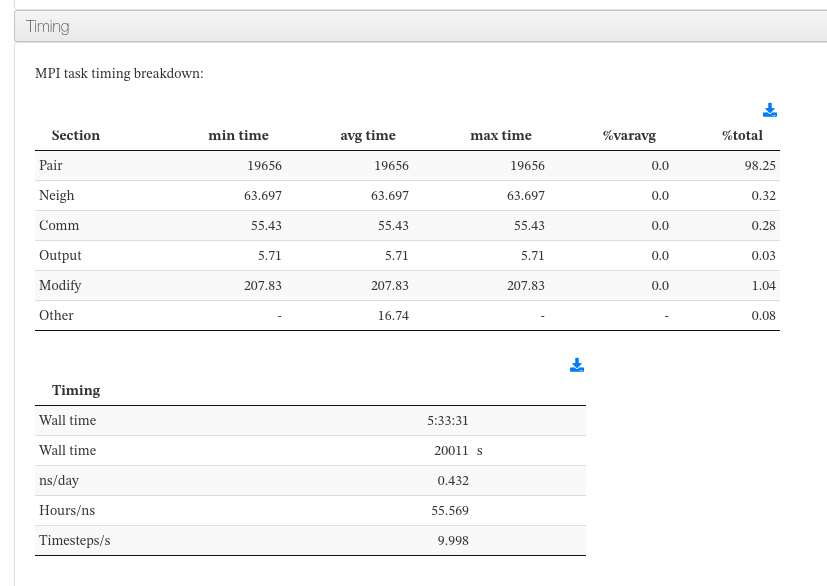
Timing
Data sources:
<module cmlx:templateRef='mpi'>
<module cmlx:templateRef='walltime'>
<module cmlx:templateRef='performance'>
[^1]: string l:getMethod string setup
$setup Refers to all defined setup parameters from <module cmlx:templateRef="setup">
<xsl:variable name="l:npt.regex">.*\s+npt.*</xsl:variable>
<xsl:variable name="l:nvt.regex">.*\s+(nvt.*|langevin.*|temp/.*)\s+.*</xsl:variable>
<xsl:variable name="l:nvt.pairstyle.regex">.*\s+(dpd/tstat.*)\s+.*</xsl:variable>
<xsl:variable name="l:nph.regex">.*\s+nph\s+.*</xsl:variable>
<xsl:variable name="l:nve.regex">.*\s+nve\s*.*</xsl:variable>
<xsl:variable name="l:minimize">Geometry optimization</xsl:variable>
<xsl:variable name="l:md">Molecular Dynamics</xsl:variable>
<xsl:variable name="l:npt">NPT</xsl:variable>
<xsl:variable name="l:nvt">NVT</xsl:variable>
<xsl:variable name="l:nph">NPH</xsl:variable>
<xsl:variable name="l:nve">NVE</xsl:variable>
<xsl:function name="l:getMethod">
<xsl:param name="setup" />
<xsl:variable name="methods">
<xsl:if test="exists(l:getParameter($setup, 'minimize'))">
<xsl:value-of select="$l:minimize"/><xsl:value-of select="'|'"/>
</xsl:if>
<xsl:for-each select="l:getParameter($setup, 'pair_style')">
<xsl:if test="matches(.,$l:nvt.pairstyle.regex)">
<xsl:value-of select="$l:nvt"/><xsl:value-of select="'|'"/>
</xsl:if>
</xsl:for-each>
<xsl:for-each select="l:getParameter($setup, 'fix')">
<xsl:choose>
<xsl:when test="matches(., $l:npt.regex)"><xsl:value-of select="$l:npt"/><xsl:value-of select="'|'"/></xsl:when>
<xsl:when test="matches(., $l:nvt.regex)"><xsl:value-of select="$l:nvt"/><xsl:value-of select="'|'"/></xsl:when>
<xsl:when test="matches(., $l:nph.regex)"><xsl:value-of select="$l:nph"/><xsl:value-of select="'|'"/></xsl:when>
<xsl:when test="matches(., $l:nve.regex)"><xsl:value-of select="$l:nve"/><xsl:value-of select="'|'"/></xsl:when>
</xsl:choose>
</xsl:for-each>
</xsl:variable>
<xsl:value-of select="distinct-values(tokenize(substring($methods, 1, string-length($methods)-1), '\|'))"/>
</xsl:function>[^2]: string l:getCouplingMethods string setup
$setup Refers to all defined setup parameters from <module cmlx:templateRef="setup">
<!-- thermostat / barostat types -->
<xsl:variable name="l:thermostat.nosehoover">Nose-Hoover</xsl:variable>
<xsl:variable name="l:thermostat.langevin">Langevin</xsl:variable>
<xsl:variable name="l:thermostat.berendsen">Berendsen</xsl:variable>
<xsl:variable name="l:thermostat.hd">Hamiltonian dynamics</xsl:variable>
<xsl:variable name="l:thermostat.ld">Langevin dynamics</xsl:variable>
<xsl:variable name="l:thermostat.rescaling">Rescaling</xsl:variable>
<xsl:variable name="l:thermostat.dpd">Dissipative particle dynamics</xsl:variable>
<xsl:variable name="l:barostat.nosehoover">Nose-Hoover</xsl:variable>
<xsl:variable name="l:barostat.berendsen">Berendsen</xsl:variable>
<!-- nvt parameters -->
<xsl:variable name="l:nvt.nosehoover.regex">.*\s+nvt.*</xsl:variable>
<xsl:variable name="l:nvt.langevin.regex">.*\s+langevin.*</xsl:variable>
<xsl:variable name="l:nvt.berendsen.regex">.*\s+temp/berendsen.*</xsl:variable>
<xsl:variable name="l:nvt.hd.regex">.*\s+temp/csvr.*</xsl:variable>
<xsl:variable name="l:nvt.ld.regex">.*\s+temp/csld.*</xsl:variable>
<xsl:variable name="l:nvt.rescaling.regex">.*\s+temp/rescale.*</xsl:variable>
<xsl:variable name="l:nvt.dpd.regex">.*\s+dpd/tstat.*</xsl:variable>
<!-- npt parameters -->
<xsl:variable name="l:npt.nosehoover.regex">.*\s+npt.*</xsl:variable>
<!-- nph parameters -->
<xsl:variable name="l:nph.nosehoover.regex">.*\s+nph.*</xsl:variable>
<xsl:variable name="l:nph.berendsen.regex">.*\s+press/berendsen.*</xsl:variable>
<xsl:function name="l:getCouplingMethods">
<xsl:param name="setup" />
<xsl:variable name="cmethods">
<xsl:for-each select="l:getParameter($setup, 'pair_style')">
<xsl:if test="matches(.,$l:nvt.pairstyle.regex)">
<xsl:value-of select="concat('(T=', $l:thermostat.dpd,')|')"/>
</xsl:if>
</xsl:for-each>
<xsl:for-each select="l:getParameter($setup, 'fix')">
<!-- nvt -->
<xsl:if test="matches(., $l:nvt.nosehoover.regex)"><xsl:value-of select="concat('(T=', $l:thermostat.nosehoover,')|')"/></xsl:if>
<xsl:if test="matches(., $l:nvt.langevin.regex)"><xsl:value-of select="concat('(T=', $l:thermostat.langevin,')|')"/></xsl:if>
<xsl:if test="matches(., $l:nvt.berendsen.regex)"><xsl:value-of select="concat('T=', $l:thermostat.berendsen,')|')"/></xsl:if>
<xsl:if test="matches(., $l:nvt.hd.regex)"><xsl:value-of select="concat('(T=', $l:thermostat.hd,')|')"/></xsl:if>
<xsl:if test="matches(., $l:nvt.ld.regex)"><xsl:value-of select="concat('(T=', $l:thermostat.ld,')|')"/></xsl:if>
<xsl:if test="matches(., $l:nvt.rescaling.regex)"><xsl:value-of select="concat('(T=', $l:thermostat.rescaling,')|')"/></xsl:if>
<xsl:if test="matches(., $l:nvt.dpd.regex)"><xsl:value-of select="concat('(T=', $l:thermostat.dpd,')|')"/></xsl:if>
<!-- npt -->
<xsl:if test="matches(., $l:npt.nosehoover.regex)"><xsl:value-of select="concat('(T=', $l:thermostat.nosehoover,', P=', $l:barostat.nosehoover,')|')"/></xsl:if>
<!-- nph -->
<xsl:if test="matches(., $l:nph.nosehoover.regex)"><xsl:value-of select="concat('(P=', $l:barostat.nosehoover,')|')"/></xsl:if>
<xsl:if test="matches(., $l:nph.berendsen.regex)"><xsl:value-of select="concat('(P=', $l:barostat.berendsen,')|')"/></xsl:if>
</xsl:for-each>
</xsl:variable>
<xsl:value-of select="distinct-values(tokenize(substring($cmethods, 1, string-length($cmethods)-1), '\|'))"/>
</xsl:function>