Molcas
Molcas
General Info
Molcas - General Info - Main fields
| Field | Source | Sample value |
|---|---|---|
| Title | Set on Browse calculation publication | Sample calculation |
| Browse Item | URL pointing Browse published item | https://rodi.urv.es:8080/browse/handle/123456789/6 |
| Program | program.header template | Molcas 8.0 - service pack 1 |
| Author | Username fullname | Doe, John |
| Formula | Atom count from final geometry | C 8 H 12 N 8 O 2 |
| Calculation type | Custom logic [^1] | Geometry optimization |
| Method | Custom logic [^2] | RASSCF RASPT2 |

Atomics and Basis Sets
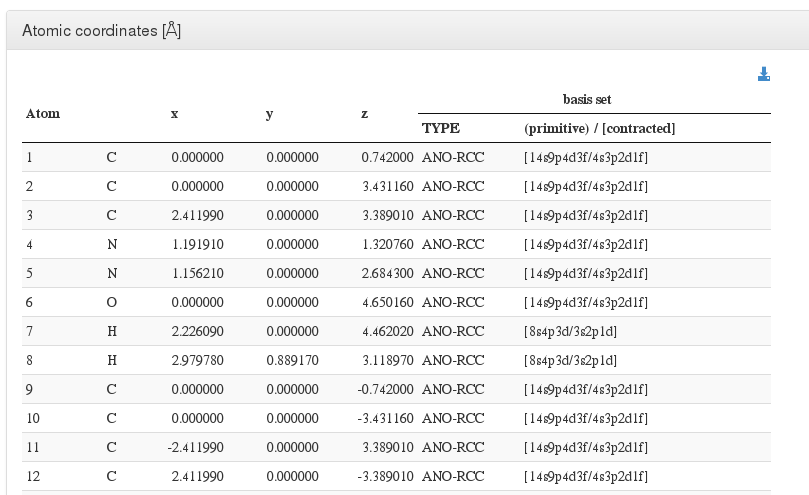
After header section, our HTML resume will output a xyz coordinates table with current molecule atoms.
For every atom, we will output it's serial number, atom type, coordinates in angstroms, basis used and contraction.
In geometry optimizations calculations, next to geometry section header there will appear the word (optimized), pointing that this geometry is the last one from all optimization steps and has converged.
If the geometry optimization did not converge, there will appear the phrase (calculation did not converge).
Molecular Info
This section captures molecule additional information not captured on previous section.
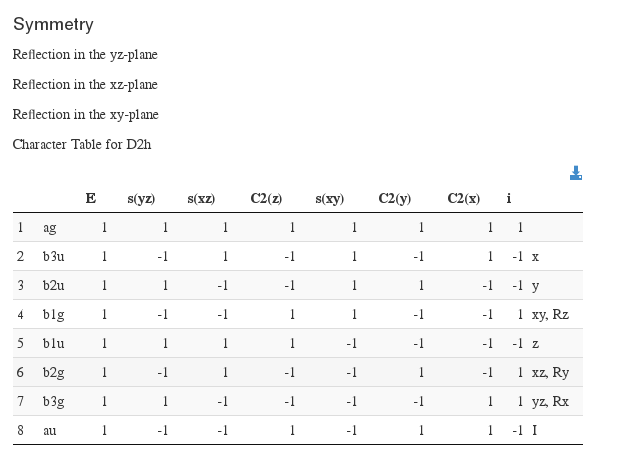
Symmetry information
Data source: <module cmlx:templateRef='symmetry'>
Molecular Info
Molecular Info - Main fields
| Field | Source | Sample value |
|---|---|---|
| Charge | Readed from "charge" scalar on molcharge line or from mulliken module | 0.000 |
| Multiplicity | Readed from scalar[m:spinquantumnum] inside wave.specs module OR from scalar[m:spin] inside scf-ksdft module | 3 |
Solvation input
Data source: <module cmlx:templateRef='pcm'>
Data source: <module cmlx:templateRef='kirkwood'>

Integrals
Data source: <module cmlx:templateRef='seward.generate'>
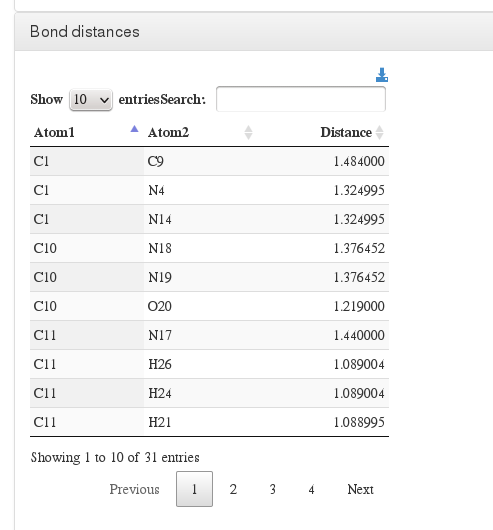
Bond distances
Data source (to read molecule and calculate bonds): <module cmlx:templateRef='coordinates'>
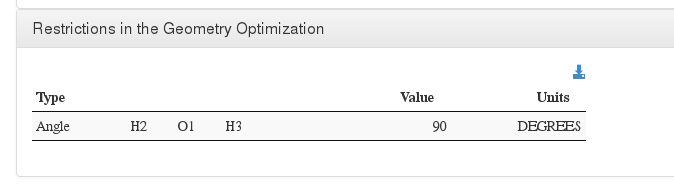
Restrictions in the Geometry Optimization
Data source: <module cmlx:templateRef='constraint'>
Modules
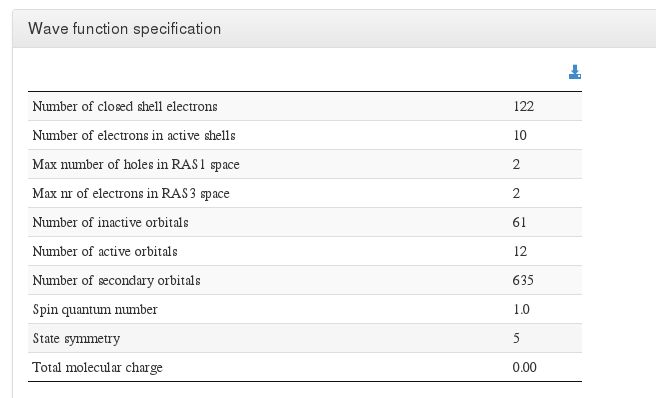
Wave function specification
Data source: <module cmlx:templateRef='wave.specs'>
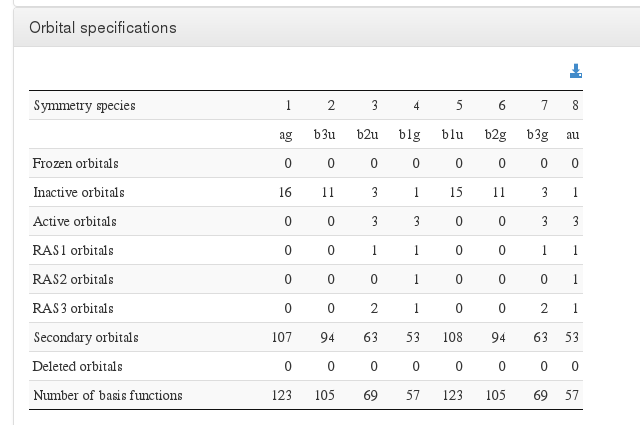
Orbital specifications
Data source: <module cmlx:templateRef='orbital.specs'>
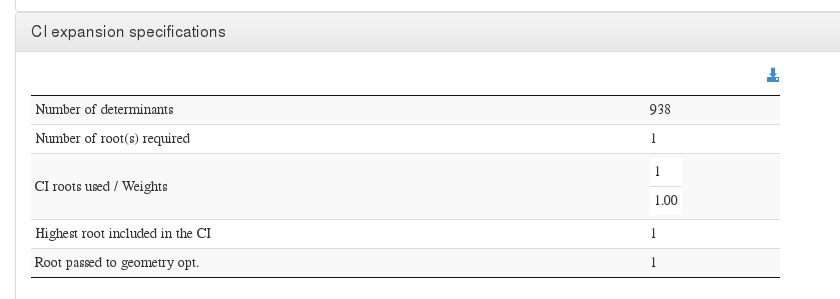
CI expansion specifications
Data source: <module cmlx:templateRef='ci.expansion'>
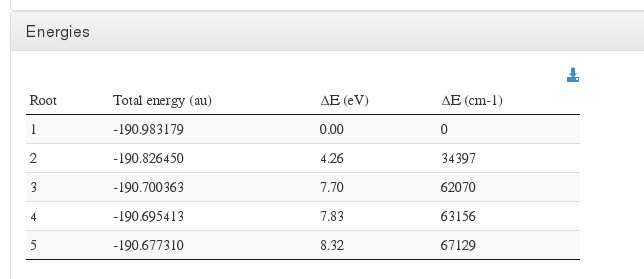
Energies
Data source: <module cmlx:templateRef='wave.printout'>
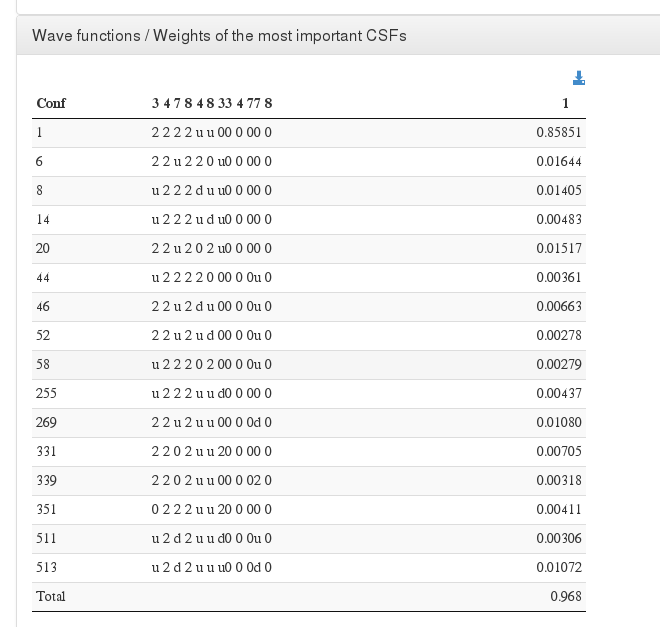
Wave functions / Weights of the most important CSFs
Data source <module cmlx:templateRef='wave.printout'>
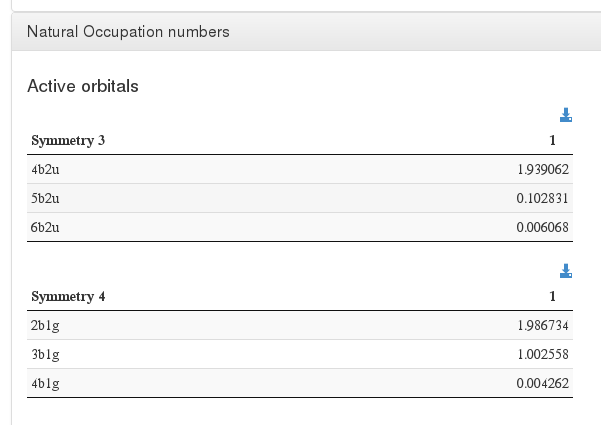
Natural Occupation numbers
Data source: <module cmlx:templateRef='wave.printout'>, module "natural"
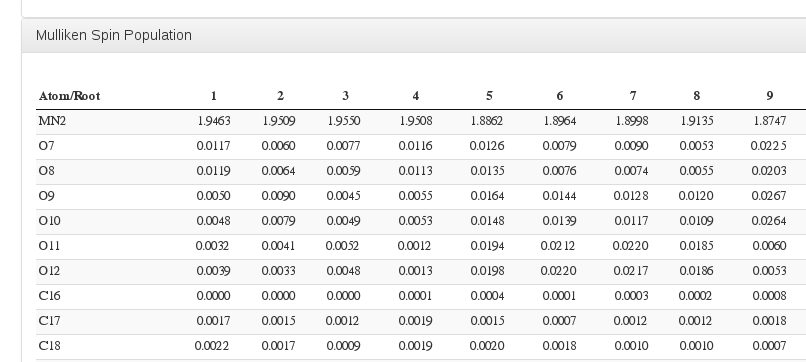
Mulliken Spin Population
Data source: <module cmlx:templateRef='mulliken'>, submodule "mulliken.spin"
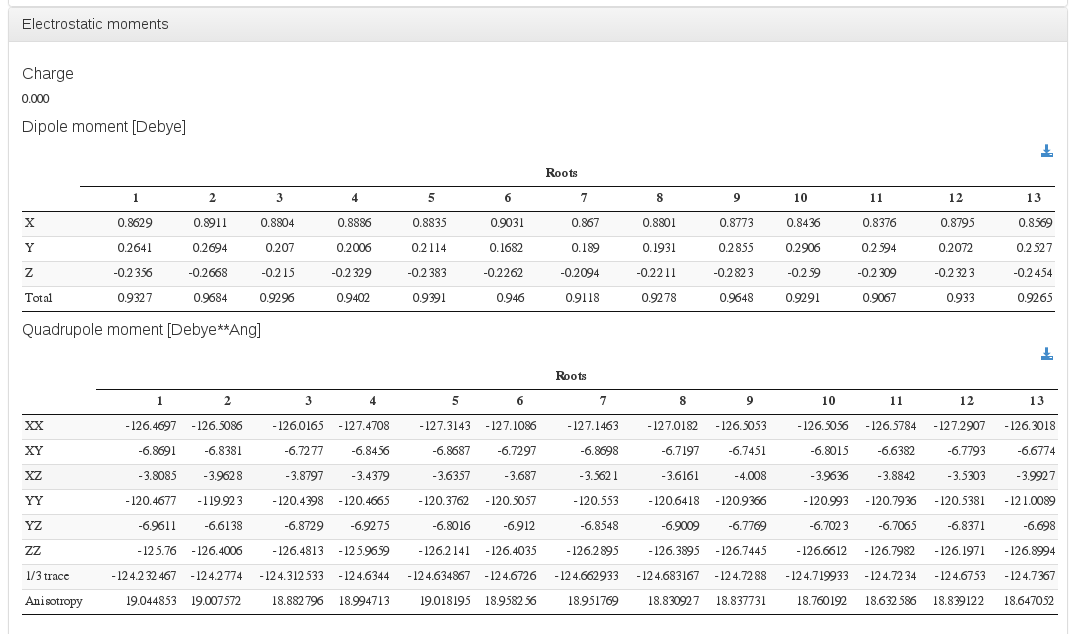
Electrostatic moments
Data source: <module cmlx:templateRef="properties">
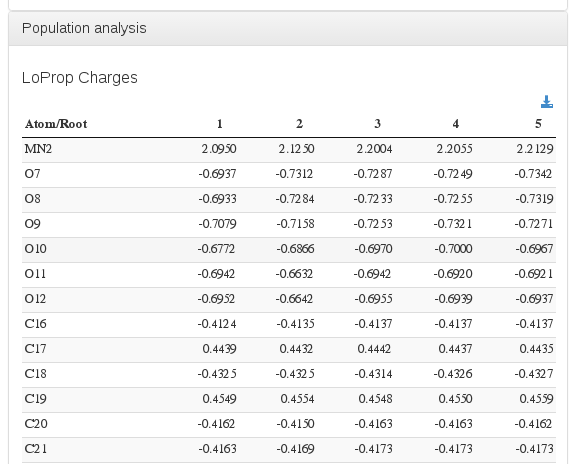
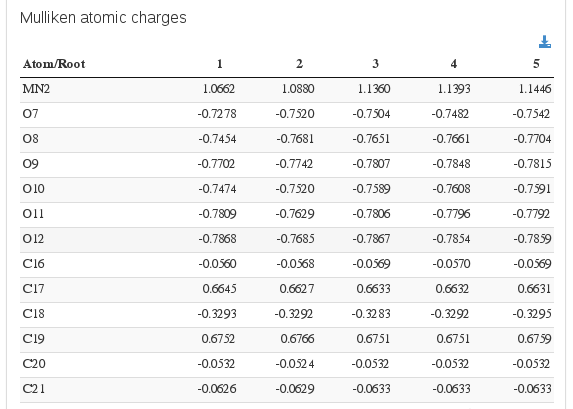
Population analysis / Mulliken atomic charges
Data source: <module cmlx:templateRef="loprop">
Data source: <module cmlx:templateRef="mulliken">
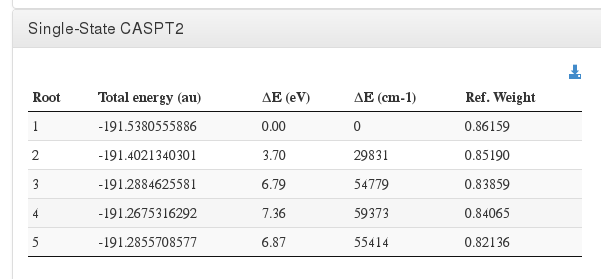
Single-State CASPT2
Data source: <module cmlx:templateRef="final.caspt2">
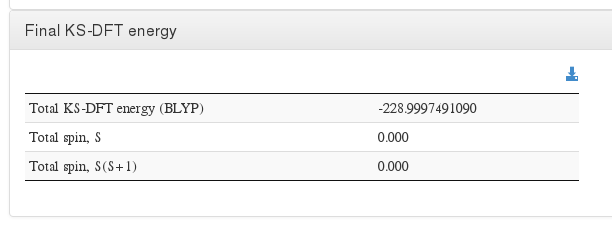
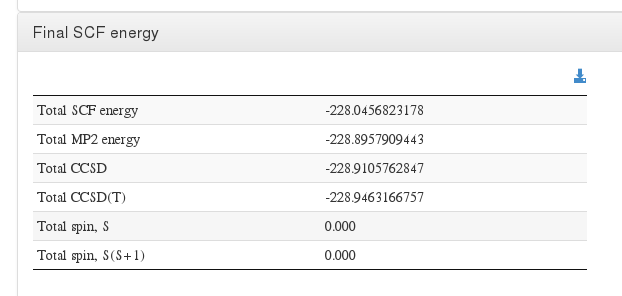
Final energy
Data source: <module cmlx:templateRef="scf-ksdft"> scalar dictRef='m:scfener'
Data source: <module cmlx:templateRef="cchc"> scalar dictRef='m:e2mp2energy'
Data source: <module cmlx:templateRef="cchc"> scalar dictRef='m:e2ccsdenergy'
Data source: <module cmlx:templateRef="ccsdt"> scalar dictRef='m:ccsdtcorrenergy'
HZERO
Data source: <module cmlx:templateRef="extras">

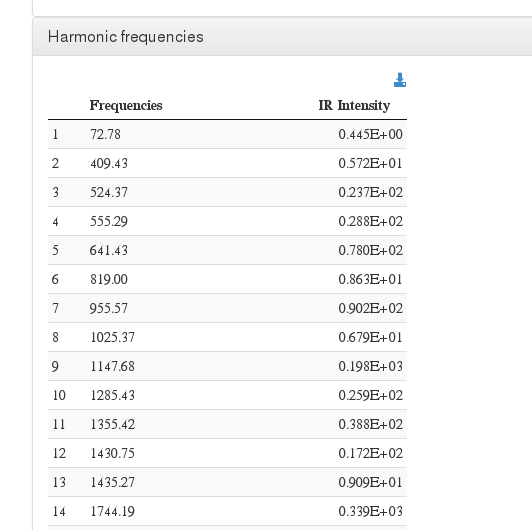
Harmonic frequencies
This module also allows displaying harmonic frequency intensities on a customizable chart.
Data source: <module cmlx:templateRef='vibrations'>
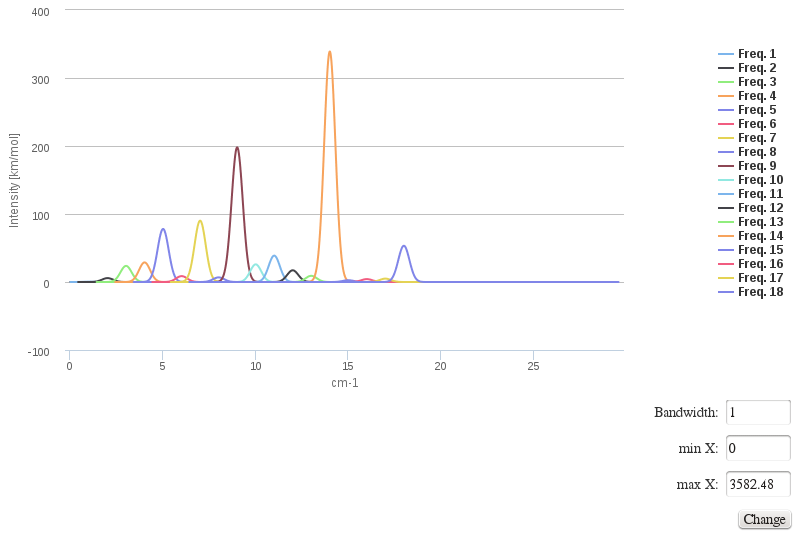
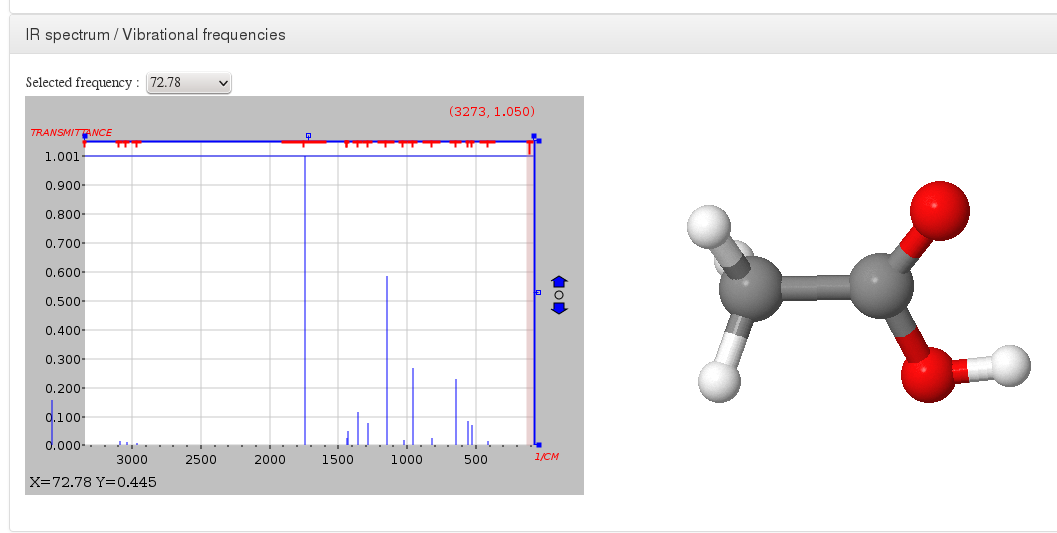
IR spectrum / Vibrational frequencies
Data source: <module cmlx:templateRef='vibrations'>
This module will display JSpecView + JSmol plugins (using javascript libraries) working together to represent molecule IR spectrum.
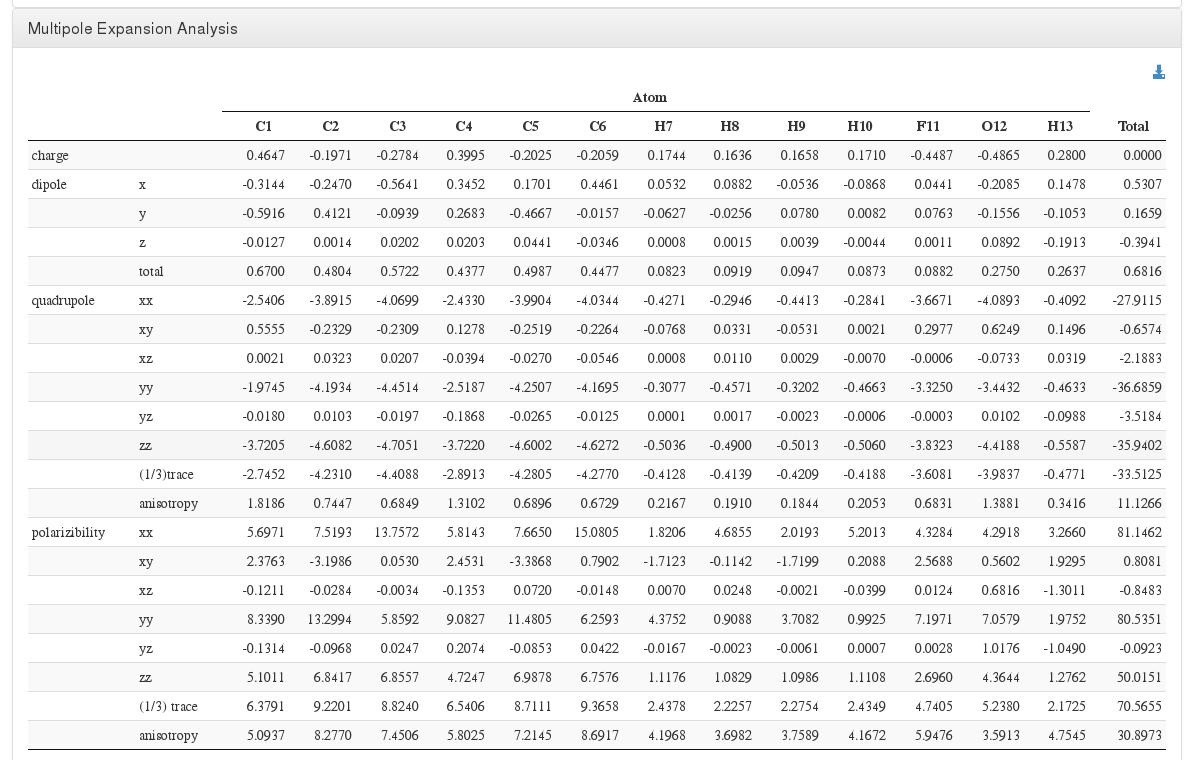
Multipole Expansion Analysis
Data source:<module cmlx:templateRef="atom.expansion">
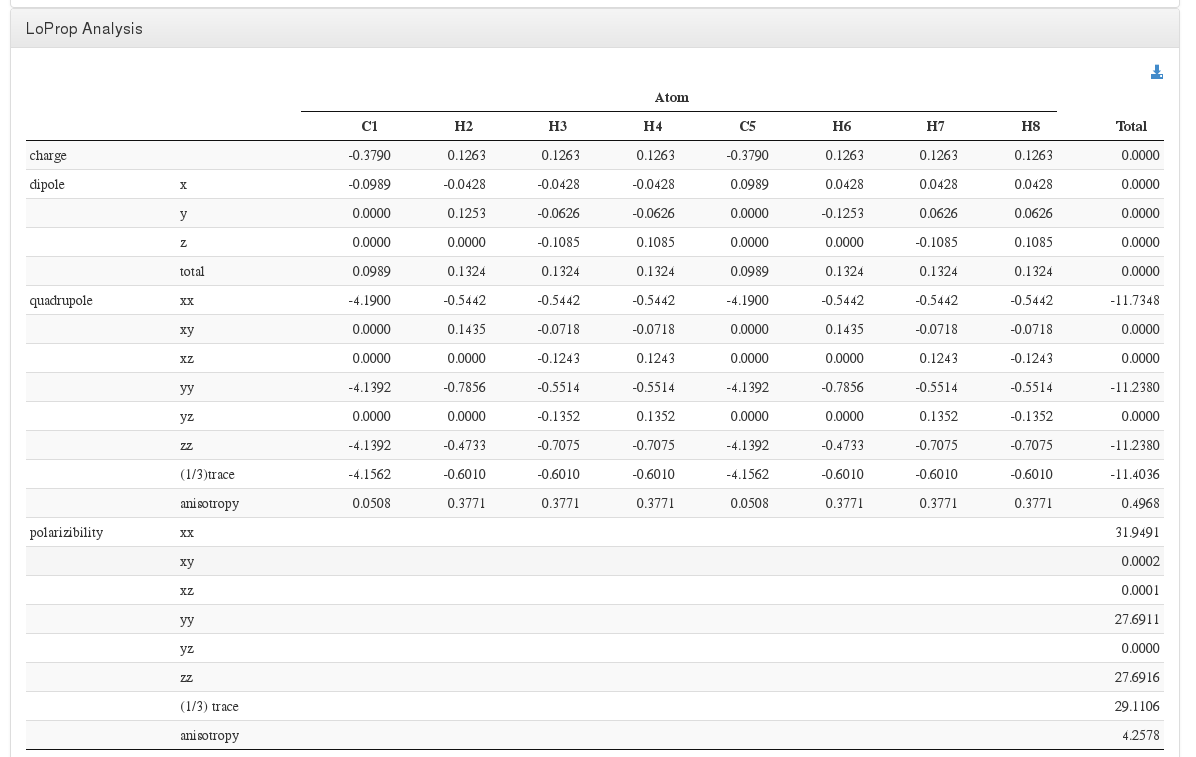
LoProp Analysis
Data source: <module cmlx:templateRef='dynamic.loprop'>
[^1]: string molcas:getCalcType boolean isRestrictedOpt boolean isOptimization boolean isTS boolean isIncomplete
$isRestrictedOpt Exists module <module cmlx:templateRef="constraint" > ?
$isOptimization Input file from <module cmlx:templateRef="molcas.input" > is setup to perform a geometry optimization?
$isTS TS keyword is defined inside <module cmlx:templateRef="molcas.input" > ?
$isIncomplete Convergence table from <module cmlx:templateRef="energy.statistics" > shows 'all converged' OR $isOptimization and $hasLastEnergySection ?
<!-- Calculation type related constants -->
<xsl:param name="isRestrictedOpt"/>
<xsl:param name="isOptimization"/>
<xsl:param name="isTS"/>
<xsl:param name="isIncomplete"/>
<xsl:choose>
<xsl:when test="$isRestrictedOpt">
<xsl:value-of select="$molcas:RestrictedGeomOpt"/>
</xsl:when>
<xsl:when test="$isOptimization">
<xsl:value-of select="$molcas:GeometryOpt"/>
</xsl:when>
<xsl:otherwise>
<xsl:value-of select="$molcas:SinglePoint"/>
</xsl:otherwise>
</xsl:choose>
<xsl:if test="$isTS">
<xsl:text> </xsl:text><xsl:value-of select="$molcas:TS"/>
</xsl:if>
<xsl:if test="$isIncomplete">
<xsl:text> </xsl:text><xsl:value-of select="$molcas:Incomplete"/>
</xsl:if>[^2]: string molcas:getMethods node* modules node ksdft node wavespecs
$modules Array with all executed module names
$ksdft Module from <module cmlx:templateRef="scf-ksdft" >
$wavespecs Entire <module cmlx:templateRef="wave.specs" > module
<xsl:param name="modules"/>
<xsl:param name="ksdft" />
<xsl:param name="wavespecs" />
<xsl:variable name="isCASSCF" select="
if(exists($wavespecs) and contains($modules,$molcas:RASSCFmodule) and ($wavespecs/cml:scalar[@dictRef='m:ras1holes'] = '0') and ($wavespecs/cml:scalar[@dictRef='m:ras3holes'] = 0)) then
true()
else
false()"/>
<xsl:variable name="isRASSCF" select="
if(contains($modules,$molcas:RASSCFmodule)) then
true()
else
false()"/>
<xsl:variable name="moduleArray">
<xsl:for-each select="$modules">
<xsl:value-of select="concat(upper-case(string(.)),'|')"/>
</xsl:for-each>
</xsl:variable>
<xsl:for-each select="distinct-values(tokenize($moduleArray/text(),'[|]+'))">
<xsl:if test="matches(.,$molcas:methodsRegex)">
<xsl:choose>
<xsl:when test="exists($ksdft) and matches(.,$molcas:SCFmodule)">
<xsl:choose>
<xsl:when test="matches($ksdft,$molcas:SCFmodule)">
<xsl:text>HF </xsl:text>
</xsl:when>
<xsl:otherwise>
<xsl:text>DFT </xsl:text>
</xsl:otherwise>
</xsl:choose>
</xsl:when>
<xsl:when test="matches(.,$molcas:RASSCFmodule)">
<xsl:choose>
<xsl:when test="$isCASSCF">
<xsl:value-of select="$molcas:CASSCFmodule"/><xsl:text> </xsl:text>
</xsl:when>
<xsl:otherwise>
<xsl:value-of select="."/><xsl:text> </xsl:text>
</xsl:otherwise>
</xsl:choose>
</xsl:when>
<xsl:when test="matches(.,$molcas:CASPT2module)">
<xsl:choose>
<xsl:when test="not($isCASSCF) and $isRASSCF">
<xsl:value-of select="$molcas:RASPT2module"/><xsl:text> </xsl:text>
</xsl:when>
<xsl:otherwise>
<xsl:value-of select="."/><xsl:text> </xsl:text>
</xsl:otherwise>
</xsl:choose>
</xsl:when>
<xsl:otherwise>
<xsl:value-of select="."/><xsl:text> </xsl:text>
</xsl:otherwise>
</xsl:choose>
</xsl:if>
</xsl:for-each>