SIESTA
SIESTA
For this specific format, the uploaded output file is the original XML generated by SIESTA.
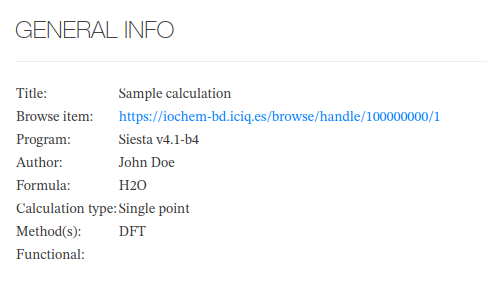
General Info
SIESTA - General Info - Main fields
| Field | Source | Sample value |
|---|---|---|
| Title | Set on Browse calculation publication | Benzene molecule |
| Browse Item | URL pointing Browse published item | https://www.iochem-bd.org/handle/10/123456 |
| Program |
| Siesta v4.1-b4 |
| Author | Username fullname | Doe, John |
| Formula | Atom count from final geometry | C6H6 |
| Calculation type | Custom logic 1 | Geometry optimization |
| Method | Method used (fixed) | DFT |
| Functional | N/A |
string
siesta:normalizeRunTypestringtype↩︎$type Value of the parameter <parameter name='MD.TypeOfRun'> <!-- Calculation type related constants --> <xsl:param name="type" as="xs:string"/> <xsl:choose> <xsl:when test="matches($type,'BROYDEN') or matches($type,'CG') or matches($type,'FIRE')"> <xsl:value-of select="'Geometry optimization'"/> </xsl:when> <xsl:when test="matches($type, 'VERLET') or matches($type, 'NOSE') or matches($type, 'PARRINELLORAHMAN') or matches($type, 'NOSEPARRINELLORAHMAN') or matches($type, 'ANNEAL') or matches($type, 'LUA')"> <xsl:value-of select="'Molecular Dynamics'"/> </xsl:when> <xsl:when test="matches($type, 'FC')"> <xsl:value-of select="'Phonon'"/> </xsl:when> <xsl:when test="matches($type, 'SINGLE-POINT')"> <xsl:value-of select="'Single point'"/> </xsl:when> <xsl:when test="matches($type, 'MASTER') or matches($type, 'FORCES')"> <xsl:value-of select="''"/> </xsl:when> <xsl:otherwise> <xsl:value-of select="''"/> </xsl:otherwise> </xsl:choose>
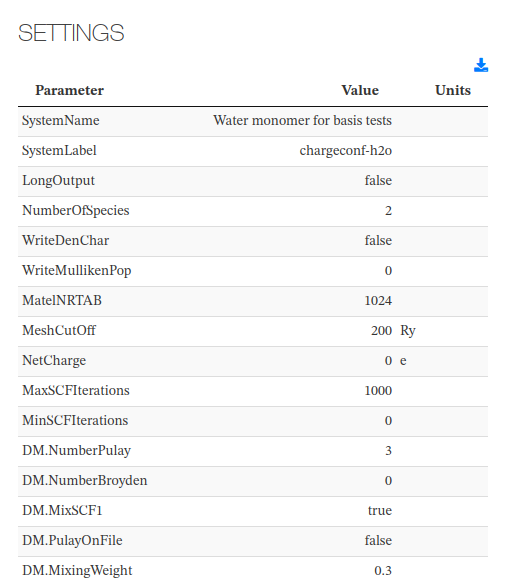
Settings
Most relevant calculation input parameters.
All fields come from the <cml><parameterList><parameter> entries.
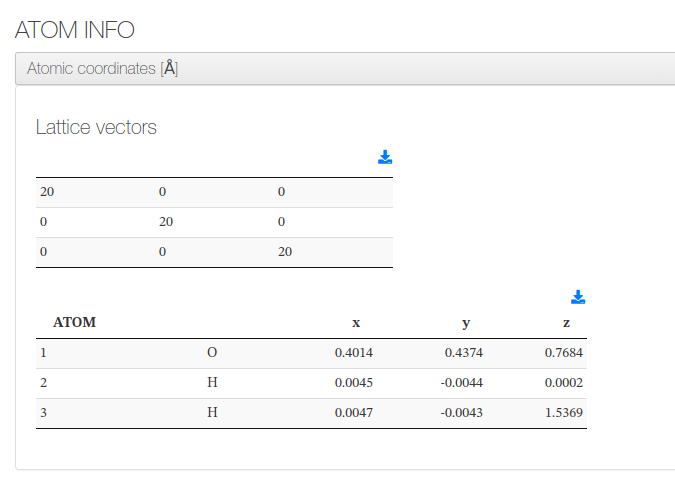
Atoms and lattice vectors
After settings section, our HTML resume will output lattice vectors and a coordinates table with molecule atoms.
Geometry is read from the final geometry at <cml><module[@title='Finalization']> or else from <cml><module[@title='Initial System']>
For every atom, we will output it's serial number, atom type, cartesian coordinates (in angstroms).
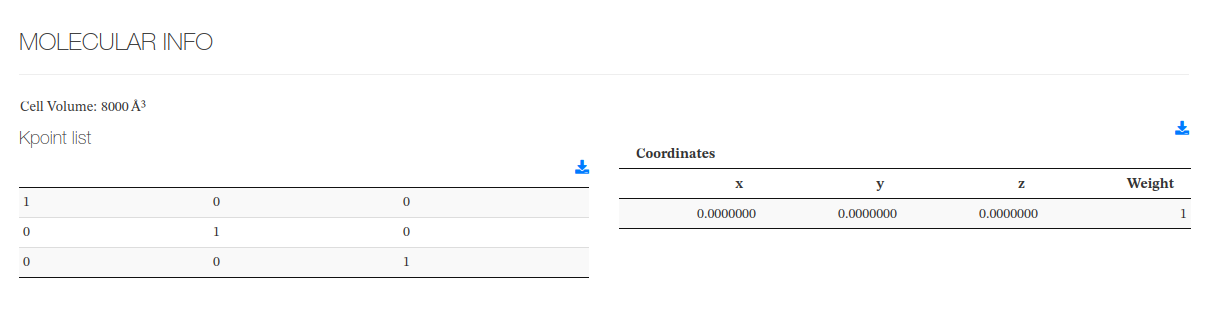
Molecular Info
This section captures molecule additional information not captured on previous section.
Volume data source: <cml><module title='Finalization'><propertyList title='Final Pressure'><property dictRef='siesta:cellvol'>
Kpoint cell data source: <cml><property dictRef='siesta:kscell'>
Kpoint list data source: <cml><propertyList title='k-points']><kpoint>
Modules
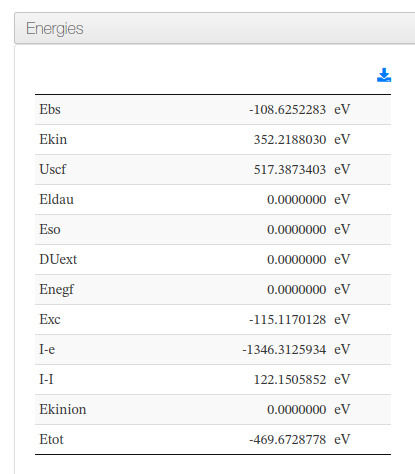
Energies
Data source: <cml><module title='Finalization'><propertyList title='Final Energy'>
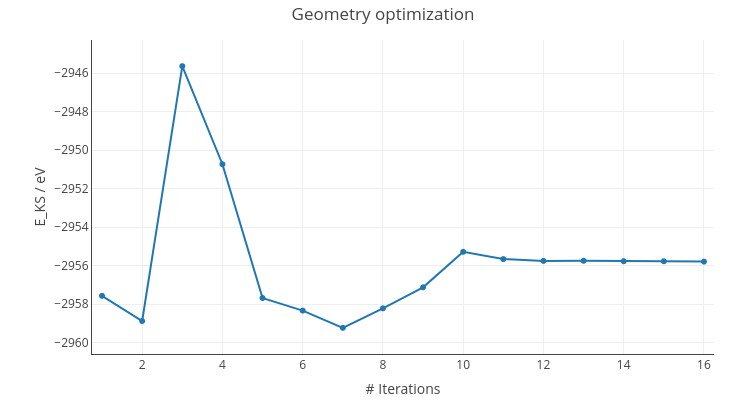
On geometry optimizations, a graph will depict Kohn-Sham energy progress.
Pressure
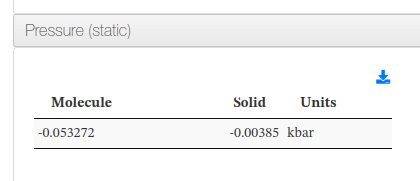
Data source: <cml><module title='Finalization'><propertyList title='Final Pressure'><property dictRef='siesta:pressSol'>
Data source: <cml><module title='Finalization'><propertyList title='Final Pressure'><property dictRef='siesta:pressMol'>
Timing
Data source <cml><metadataList><metadata name='siesta:Mode'>
Data source <cml><metadataList><metadata name='siesta:Nodes'>
Data source <cml><metadataList><metadata name='siesta:StartTime'>
Data source <cml><metadataList><metadata name='siesta:EndTime'>

